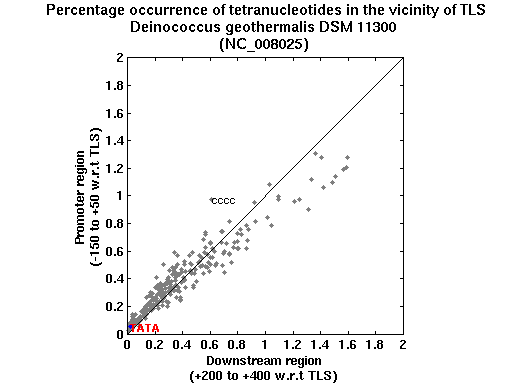
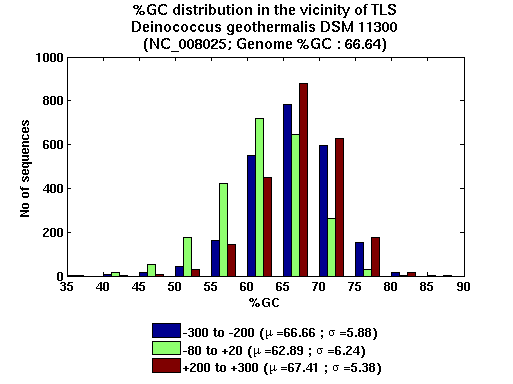
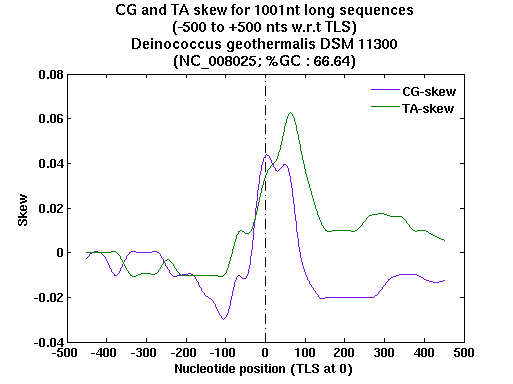
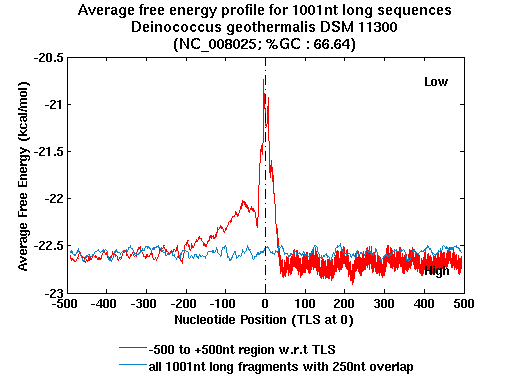
| Gene ID |
Gene name |
Left end
position |
Right end
position |
Length |
Transcription
direction |
Gene Product |
| Dgeo_0001 | Dgeo_0001 | 222 | 1634 | 1413 | + | PROTEIN:chromosomal replication initiator protein |
| Dgeo_0003 | Dgeo_0003 | 2025 | 3107 | 1083 | + | PROTEIN:DNA polymerase III, beta subunit |
| Dgeo_0004 | Dgeo_0004 | 3245 | 4513 | 1269 | + | PROTEIN:phosphopyruvate hydratase |
| Dgeo_0005 | Dgeo_0005 | 4600 | 6048 | 1449 | + | PROTEIN:pyruvate kinase |
| Dgeo_0006 | Dgeo_0006 | 6295 | 7536 | 1242 | - | PROTEIN:tyrosyl-tRNA synthetase |
| Dgeo_0007 | Dgeo_0007 | 7665 | 8285 | 621 | + | PROTEIN:MOSC domain-containing protein |
| Dgeo_0008 | Dgeo_0008 | 8301 | 8852 | 552 | + | PROTEIN:hypothetical protein |
| Dgeo_0009 | Dgeo_0009 | 8833 | 9510 | 678 | + | PROTEIN:hypothetical protein |
| Dgeo_0010 | Dgeo_0010 | 9612 | 11432 | 1821 | - | PROTEIN:glucosamine--fructose-6-phosphate aminotra |
| Dgeo_0011 | Dgeo_0011 | 11803 | 13008 | 1206 | - | PROTEIN:methionine gamma-lyase |
| Dgeo_0012 | Dgeo_0012 | 13036 | 13686 | 651 | - | PROTEIN:hypothetical protein |
| Dgeo_0013 | rpmF | 13862 | 14044 | 183 | + | PROTEIN:50S ribosomal protein L32 |
| Dgeo_0014 | Dgeo_0014 | 14304 | 15089 | 786 | + | PROTEIN:response regulator receiver/SARP domain-co |
| Dgeo_0015 | Dgeo_0015 | 15138 | 15407 | 270 | + | PROTEIN:hypothetical protein |
| Dgeo_0016 | Dgeo_0016 | 15559 | 16119 | 561 | + | PROTEIN:cytochrome c oxidase, subunit II |
| Dgeo_0017 | Dgeo_0017 | 16130 | 17893 | 1764 | + | PROTEIN:cytochrome c oxidase, subunit I |
| Dgeo_0018 | Dgeo_0018 | 17893 | 18309 | 417 | + | PROTEIN:cytochrome c, class I |
| Dgeo_0019 | Dgeo_0019 | 18407 | 19438 | 1032 | + | PROTEIN:A/G-specific adenine glycosylase |
| Dgeo_0020 | Dgeo_0020 | 19494 | 20324 | 831 | + | PROTEIN:undecaprenyl diphosphate synthase |
| Dgeo_0021 | Dgeo_0021 | 20373 | 22334 | 1962 | + | PROTEIN:HRDC |
| Dgeo_0022 | Dgeo_0022 | 22331 | 22666 | 336 | - | PROTEIN:hypothetical protein |
| Dgeo_0023 | Dgeo_0023 | 22666 | 23487 | 822 | - | PROTEIN:ABC transporter related |
| Dgeo_0024 | Dgeo_0024 | 23480 | 24499 | 1020 | - | PROTEIN:transport system permease protein |
| Dgeo_0025 | Dgeo_0025 | 24503 | 25372 | 870 | - | PROTEIN:periplasmic binding protein |
| Dgeo_0026 | Dgeo_0026 | 25679 | 26323 | 645 | - | PROTEIN:phospholipid/glycerol acyltransferase |
| Dgeo_0027 | Dgeo_0027 | 26392 | 26643 | 252 | - | PROTEIN:hypothetical protein |
| Dgeo_0029 | Dgeo_0029 | 27484 | 27840 | 357 | - | PROTEIN:hypothetical protein |
| Dgeo_0030 | Dgeo_0030 | 27906 | 28526 | 621 | - | PROTEIN:hypothetical protein |
| Dgeo_0031 | Dgeo_0031 | 28523 | 29392 | 870 | - | PROTEIN:hypothetical protein |
| Dgeo_0032 | Dgeo_0032 | 29584 | 30993 | 1410 | + | PROTEIN:ribonuclease II |
| Dgeo_0033 | Dgeo_0033 | 31038 | 31328 | 291 | + | PROTEIN:hypothetical protein |
| Dgeo_0034 | Dgeo_0034 | 31472 | 32503 | 1032 | + | PROTEIN:peptidyl-arginine deiminase |
| Dgeo_0035 | Dgeo_0035 | 32488 | 33663 | 1176 | + | PROTEIN:hypothetical protein |
| Dgeo_0036 | Dgeo_0036 | 33677 | 34561 | 885 | + | PROTEIN:Nitrilase/cyanide hydratase and apolipopro |
| Dgeo_0037 | Dgeo_0037 | 34598 | 35398 | 801 | - | PROTEIN:tRNA pseudouridine synthase A |
| Dgeo_0038 | Dgeo_0038 | 35401 | 36393 | 993 | + | PROTEIN:peptidase M23B |
| Dgeo_0039 | Dgeo_0039 | 36398 | 36625 | 228 | + | PROTEIN:hypothetical protein |
| Dgeo_0040 | Dgeo_0040 | 36626 | 37411 | 786 | - | PROTEIN:stationary-phase survival protein SurE |
| Dgeo_0041 | Dgeo_0041 | 37477 | 38694 | 1218 | - | PROTEIN:alcohol dehydrogenase GroES-like protein |
| Dgeo_0042 | Dgeo_0042 | 38691 | 39425 | 735 | - | PROTEIN:cyclase/dehydrase |
| Dgeo_0043 | Dgeo_0043 | 39684 | 40118 | 435 | + | PROTEIN:response regulator receiver protein |
| Dgeo_0044 | Dgeo_0044 | 40286 | 40609 | 324 | + | PROTEIN:multisubunit Na+/H+ antiporter MnhE subuni |
| Dgeo_0045 | Dgeo_0045 | 40596 | 41294 | 699 | - | PROTEIN:hypothetical protein |
| Dgeo_0046 | Dgeo_0046 | 41414 | 42151 | 738 | + | PROTEIN:polysaccharide deacetylase |
| Dgeo_0047 | Dgeo_0047 | 42215 | 42886 | 672 | - | PROTEIN:hypothetical protein |
| Dgeo_0048 | Dgeo_0048 | 43033 | 44091 | 1059 | + | PROTEIN:permease YjgP/YjgQ |
| Dgeo_0049 | Dgeo_0049 | 44102 | 45205 | 1104 | + | PROTEIN:permease YjgP/YjgQ |
| Dgeo_0050 | Dgeo_0050 | 45266 | 46681 | 1416 | + | PROTEIN:cyclopropane-fatty-acyl-phospholipid synth |
| Dgeo_0051 | Dgeo_0051 | 46701 | 48812 | 2112 | - | PROTEIN:phosphate acetyltransferase |
| Dgeo_0052 | Dgeo_0052 | 48809 | 49996 | 1188 | - | PROTEIN:acetate kinase |
| Dgeo_0053 | Dgeo_0053 | 50064 | 50513 | 450 | - | PROTEIN:hypothetical protein |
| Dgeo_0054 | Dgeo_0054 | 50524 | 52458 | 1935 | - | PROTEIN:hypothetical protein |
| Dgeo_0055 | Dgeo_0055 | 52487 | 54076 | 1590 | - | PROTEIN:hypothetical protein |
| Dgeo_0056 | Dgeo_0056 | 54073 | 54783 | 711 | - | PROTEIN:roadblock/LC7 domain-contain protein |
| Dgeo_0057 | Dgeo_0057 | 54859 | 56049 | 1191 | + | PROTEIN:pyridoxal phosphate-dependent acyltransfer |
| Dgeo_0058 | Dgeo_0058 | 56125 | 56844 | 720 | + | PROTEIN:ubiquinone/menaquinone biosynthesis methyl |
| Dgeo_0059 | Dgeo_0059 | 57092 | 58465 | 1374 | + | PROTEIN:Alpha,alpha-trehalose-phosphate synthase ( |
| Dgeo_0060 | Dgeo_0060 | 58462 | 59178 | 717 | + | PROTEIN:HAD family hydrolase |
| Dgeo_0061 | Dgeo_0061 | 59257 | 59499 | 243 | + | PROTEIN:hypothetical protein |
| Dgeo_0062 | Dgeo_0062 | 59640 | 60263 | 624 | + | PROTEIN:phosphoglycerate mutase |
| Dgeo_0063 | Dgeo_0063 | 60394 | 61812 | 1419 | - | PROTEIN:amidophosphoribosyltransferase |
| Dgeo_0064 | Dgeo_0064 | 61809 | 64049 | 2241 | - | PROTEIN:phosphoribosylformylglycinamidine synthase |
| Dgeo_0065 | Dgeo_0065 | 64046 | 64714 | 669 | - | PROTEIN:phosphoribosylformylglycinamidine synthase |
| Dgeo_0066 | Dgeo_0066 | 64711 | 64968 | 258 | - | PROTEIN:phosphoribosylformylglycinamidine syntheta |
| Dgeo_0067 | Dgeo_0067 | 65028 | 65753 | 726 | - | PROTEIN:phosphoribosylaminoimidazole-succinocarbox |
| Dgeo_0068 | Dgeo_0068 | 66048 | 66899 | 852 | - | PROTEIN:HemK family modification methylase |
| Dgeo_0069 | Dgeo_0069 | 66937 | 67527 | 591 | - | PROTEIN:single-stranded nucleic acid binding R3H |
| Dgeo_0070 | Dgeo_0070 | 67615 | 68199 | 585 | - | PROTEIN:peptidylprolyl isomerase |
| Dgeo_0071 | Dgeo_0071 | 68257 | 69522 | 1266 | - | PROTEIN:peptidase M29, aminopeptidase II |
| Dgeo_0072 | Dgeo_0072 | 69773 | 70234 | 462 | - | PROTEIN:tRNA/rRNA methyltransferase (SpoU) |
| Dgeo_0073 | Dgeo_0073 | 70231 | 70722 | 492 | - | PROTEIN:2-C-methyl-D-erythritol 2,4-cyclodiphospha |
| Dgeo_0074 | Dgeo_0074 | 70807 | 71430 | 624 | - | PROTEIN:hypothetical protein |
| Dgeo_0075 | Dgeo_0075 | 71427 | 72053 | 627 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_0076 | Dgeo_0076 | 72291 | 72848 | 558 | - | PROTEIN:DinB |
| Dgeo_0077 | Dgeo_0077 | 72893 | 73894 | 1002 | - | PROTEIN:dihydrouridine synthase, DuS |
| Dgeo_0078 | Dgeo_0078 | 74053 | 74559 | 507 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0079 | Dgeo_0079 | 74592 | 75341 | 750 | + | PROTEIN:acetylglutamate kinase |
| Dgeo_0080 | Dgeo_0080 | 75346 | 75954 | 609 | - | PROTEIN:ribosomal protein N-acetyltransferase, put |
| Dgeo_0081 | Dgeo_0081 | 75968 | 77083 | 1116 | - | PROTEIN:phosphoribosylaminoimidazole carboxylase, |
| Dgeo_0082 | Dgeo_0082 | 77080 | 77613 | 534 | - | PROTEIN:phosphoribosylaminoimidazole carboxylase, |
| Dgeo_0083 | Dgeo_0083 | 77636 | 78850 | 1215 | + | PROTEIN:FolC bifunctional protein |
| Dgeo_R0001 | Dgeo_R0001 | 78888 | 78963 | 76 | + | RNA:Lys tRNA |
| Dgeo_0084 | Dgeo_0084 | 79358 | 79795 | 438 | + | PROTEIN:hypothetical protein |
| Dgeo_0085 | Dgeo_0085 | 80110 | 80535 | 426 | - | PROTEIN:hypothetical protein |
| Dgeo_0086 | Dgeo_0086 | 80725 | 81318 | 594 | + | PROTEIN:hypothetical protein |
| Dgeo_0087 | Dgeo_0087 | 81377 | 82045 | 669 | + | PROTEIN:two component transcriptional regulator |
| Dgeo_0088 | Dgeo_0088 | 82042 | 83121 | 1080 | + | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_0089 | Dgeo_0089 | 83147 | 85648 | 2502 | - | PROTEIN:ATPase, P type cation/copper-transporter |
| Dgeo_0090 | Dgeo_0090 | 85816 | 86034 | 219 | + | PROTEIN:heavy metal transport/detoxification prote |
| Dgeo_0091 | Dgeo_0091 | 86034 | 86378 | 345 | + | PROTEIN:hypothetical protein |
| Dgeo_0092 | Dgeo_0092 | 86421 | 87728 | 1308 | - | PROTEIN:hypothetical protein |
| Dgeo_0093 | Dgeo_0093 | 87831 | 88760 | 930 | + | PROTEIN:porphobilinogen deaminase |
| Dgeo_0094 | Dgeo_0094 | 88951 | 89916 | 966 | + | PROTEIN:asparaginase/glutaminase |
| Dgeo_0095 | Dgeo_0095 | 89945 | 90562 | 618 | - | PROTEIN:DedA family protein |
| Dgeo_0096 | Dgeo_0096 | 90628 | 91344 | 717 | + | PROTEIN:HAD family hydrolase |
| Dgeo_0097 | Dgeo_0097 | 91376 | 92311 | 936 | - | PROTEIN:twin-arginine translocation pathway signal |
| Dgeo_0098 | Dgeo_0098 | 92543 | 93220 | 678 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0099 | Dgeo_0099 | 93423 | 94889 | 1467 | - | PROTEIN:UDP-N-acetylmuramyl-tripeptide synthetases |
| Dgeo_0100 | Dgeo_0100 | 94990 | 95373 | 384 | + | PROTEIN:ArsR family transcriptional regulator |
| Dgeo_0101 | Dgeo_0101 | 95398 | 96021 | 624 | - | PROTEIN:deoxynucleoside kinase |
| Dgeo_0102 | Dgeo_0102 | 96083 | 96700 | 618 | - | PROTEIN:deoxynucleoside kinase |
| Dgeo_0103 | Dgeo_0103 | 96805 | 97908 | 1104 | - | PROTEIN:peptidase S1 and S6, chymotrypsin/Hap |
| Dgeo_0104 | Dgeo_0104 | 97905 | 98114 | 210 | - | PROTEIN:hypothetical protein |
| Dgeo_0105 | Dgeo_0105 | 98248 | 100086 | 1839 | - | PROTEIN:acyl-CoA dehydrogenase-like protein |
| Dgeo_0106 | Dgeo_0106 | 100320 | 101084 | 765 | + | PROTEIN:Crp/FNR family transcriptional regulator |
| Dgeo_0107 | Dgeo_0107 | 101097 | 101747 | 651 | - | PROTEIN:HhH-GPD |
| Dgeo_0108 | Dgeo_0108 | 101875 | 102336 | 462 | + | PROTEIN:hypothetical protein |
| Dgeo_0109 | Dgeo_0109 | 102407 | 103123 | 717 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0110 | Dgeo_0110 | 103120 | 103530 | 411 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_0111 | Dgeo_0111 | 103615 | 105096 | 1482 | - | PROTEIN:ABC transporter related |
| Dgeo_0112 | Dgeo_0112 | 105107 | 106003 | 897 | - | PROTEIN:hypothetical protein |
| Dgeo_0113 | Dgeo_0113 | 106170 | 106475 | 306 | + | PROTEIN:hypothetical protein |
| Dgeo_0114 | Dgeo_0114 | 106584 | 107261 | 678 | + | PROTEIN:cyclase/dehydrase |
| Dgeo_0115 | Dgeo_0115 | 107434 | 107910 | 477 | - | PROTEIN:hypothetical protein |
| Dgeo_0116 | Dgeo_0116 | 108007 | 108291 | 285 | - | PROTEIN:hypothetical protein |
| Dgeo_0117 | Dgeo_0117 | 108295 | 108921 | 627 | - | PROTEIN:TetR family transcriptional regulator |
| Dgeo_0118 | Dgeo_0118 | 109033 | 110124 | 1092 | + | PROTEIN:aminotransferase, class I and II |
| Dgeo_0119 | Dgeo_0119 | 110183 | 110482 | 300 | + | PROTEIN:hypothetical protein |
| Dgeo_0120 | Dgeo_0120 | 110479 | 111642 | 1164 | - | PROTEIN:chromate transporter |
| Dgeo_0121 | Dgeo_0121 | 111733 | 112827 | 1095 | - | PROTEIN:fatty acid desaturase, type 2 |
| Dgeo_0122 | Dgeo_0122 | 113033 | 113488 | 456 | + | PROTEIN:redoxin |
| Dgeo_0123 | Dgeo_0123 | 113498 | 113932 | 435 | - | PROTEIN:hypothetical protein |
| Dgeo_0124 | Dgeo_0124 | 114089 | 114976 | 888 | + | PROTEIN:periplasmic binding protein |
| Dgeo_0125 | Dgeo_0125 | 114973 | 116043 | 1071 | + | PROTEIN:transport system permease protein |
| Dgeo_0126 | Dgeo_0126 | 116052 | 116885 | 834 | + | PROTEIN:hemin importer ATP-binding subunit |
| Dgeo_0127 | Dgeo_0127 | 116943 | 117743 | 801 | + | PROTEIN:siderophore-interacting protein |
| Dgeo_0128 | Dgeo_0128 | 117762 | 118406 | 645 | - | PROTEIN:phosphate uptake regulator, PhoU |
| Dgeo_0129 | Dgeo_0129 | 118468 | 119442 | 975 | - | PROTEIN:histidine kinase |
| Dgeo_0130 | Dgeo_0130 | 119442 | 120131 | 690 | - | PROTEIN:two component transcriptional regulator |
| Dgeo_0131 | Dgeo_0131 | 120210 | 120596 | 387 | - | PROTEIN:Iojap-related protein |
| Dgeo_0132 | Dgeo_0132 | 120619 | 121899 | 1281 | - | PROTEIN:cell envelope-related transcriptional atte |
| Dgeo_0133 | Dgeo_0133 | 121896 | 122510 | 615 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_0134 | Dgeo_0134 | 122611 | 123594 | 984 | - | PROTEIN:YbbR-like protein |
| Dgeo_0135 | Dgeo_0135 | 123591 | 124427 | 837 | - | PROTEIN:hypothetical protein |
| Dgeo_0136 | Dgeo_0136 | 124577 | 125290 | 714 | + | PROTEIN:beta-lactamase-like protein |
| Dgeo_0137 | Dgeo_0137 | 125304 | 126044 | 741 | - | PROTEIN:hypothetical protein |
| Dgeo_0138 | pyrE | 126133 | 126732 | 600 | + | PROTEIN:orotate phosphoribosyltransferase |
| Dgeo_0139 | Dgeo_0139 | 126791 | 128068 | 1278 | - | PROTEIN:2-oxoglutarate dehydrogenase, E2 component |
| Dgeo_0140 | kgd | 128181 | 131141 | 2961 | - | PROTEIN:alpha-ketoglutarate decarboxylase |
| Dgeo_0141 | Dgeo_0141 | 131393 | 132322 | 930 | + | PROTEIN:transport-associated |
| Dgeo_0142 | Dgeo_0142 | 132456 | 132731 | 276 | + | PROTEIN:hypothetical protein |
| Dgeo_0143 | Dgeo_0143 | 132759 | 133982 | 1224 | - | PROTEIN:cytochrome P450 |
| Dgeo_0144 | Dgeo_0144 | 134376 | 135275 | 900 | + | PROTEIN:pyridoxamine kinase |
| Dgeo_0145 | Dgeo_0145 | 135327 | 136070 | 744 | + | PROTEIN:enoyl-CoA hydratase/isomerase |
| Dgeo_0146 | Dgeo_0146 | 136123 | 136956 | 834 | + | PROTEIN:short chain dehydrogenase |
| Dgeo_0147 | Dgeo_0147 | 136990 | 138411 | 1422 | + | PROTEIN:peptidase S13, D-Ala-D-Ala carboxypeptidas |
| Dgeo_0148 | xseA | 138469 | 139710 | 1242 | - | PROTEIN:exodeoxyribonuclease VII large subunit |
| Dgeo_0149 | Dgeo_0149 | 139707 | 140087 | 381 | - | PROTEIN:crcB protein |
| Dgeo_0150 | Dgeo_0150 | 140133 | 141170 | 1038 | + | PROTEIN:Mg2+ transporter protein, CorA-like protei |
| Dgeo_0151 | Dgeo_0151 | 141148 | 141978 | 831 | - | PROTEIN:TatD-related deoxyribonuclease |
| Dgeo_0152 | Dgeo_0152 | 142122 | 142757 | 636 | + | PROTEIN:hypothetical protein |
| Dgeo_0153 | Dgeo_0153 | 142754 | 143647 | 894 | + | PROTEIN:OstA-like protein protein |
| Dgeo_0154 | Dgeo_0154 | 143657 | 144646 | 990 | + | PROTEIN:OstA-like protein protein |
| Dgeo_0155 | Dgeo_0155 | 144734 | 145267 | 534 | - | PROTEIN:hypothetical protein |
| Dgeo_0156 | Dgeo_0156 | 145359 | 146816 | 1458 | + | PROTEIN:beta-lactamase-like protein |
| Dgeo_0157 | Dgeo_0157 | 146843 | 147409 | 567 | - | PROTEIN:exonuclease, RNase T and DNA polymerase II |
| Dgeo_0158 | Dgeo_0158 | 147479 | 147976 | 498 | + | PROTEIN:hypothetical protein |
| Dgeo_0159 | Dgeo_0159 | 147985 | 148863 | 879 | - | PROTEIN:3-mercaptopyruvate sulfurtransferase |
| Dgeo_0160 | Dgeo_0160 | 149149 | 150003 | 855 | + | PROTEIN:aldo/keto reductase |
| Dgeo_0161 | Dgeo_0161 | 149975 | 150250 | 276 | - | PROTEIN:hypothetical protein |
| Dgeo_0162 | Dgeo_0162 | 150443 | 152131 | 1689 | - | PROTEIN:hypothetical protein |
| Dgeo_0163 | rplI | 152455 | 152895 | 441 | - | PROTEIN:50S ribosomal protein L9 |
| Dgeo_0164 | rpsR | 152907 | 153182 | 276 | - | PROTEIN:30S ribosomal protein S18 |
| Dgeo_0165 | Dgeo_0165 | 153202 | 154107 | 906 | - | PROTEIN:single-strand binding protein |
| Dgeo_0166 | rpsF | 154236 | 154544 | 309 | - | PROTEIN:30S ribosomal protein S6 |
| Dgeo_0167 | Dgeo_0167 | 154686 | 154880 | 195 | + | PROTEIN:hypothetical protein |
| Dgeo_0168 | Dgeo_0168 | 154899 | 155987 | 1089 | - | PROTEIN:serine/threonine protein kinase |
| Dgeo_0169 | Dgeo_0169 | 156077 | 157219 | 1143 | - | PROTEIN:peptide chain release factor 2 |
| Dgeo_R0002 | Dgeo_R0002 | 157596 | 159088 | 1493 | + | RNA:16S ribosomal RNA |
| Dgeo_0170 | Dgeo_0170 | 159185 | 159688 | 504 | - | PROTEIN:CarD family transcriptional regulator |
| Dgeo_0171 | Dgeo_0171 | 159941 | 160690 | 750 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0172 | Dgeo_0172 | 160687 | 161619 | 933 | - | PROTEIN:ABC transporter related |
| Dgeo_0173 | Dgeo_0173 | 161681 | 162874 | 1194 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0174 | Dgeo_0174 | 162924 | 163826 | 903 | - | PROTEIN:substrate-binding region of ABC-type glyci |
| Dgeo_0175 | Dgeo_0175 | 163941 | 164291 | 351 | - | PROTEIN:histone-like protein DNA-binding protein |
| Dgeo_0176 | Dgeo_0176 | 164378 | 165118 | 741 | - | PROTEIN:UBA/THIF-type NAD/FAD binding fold |
| Dgeo_0177 | Dgeo_0177 | 165138 | 165728 | 591 | + | PROTEIN:hypothetical protein |
| Dgeo_0178 | Dgeo_0178 | 165725 | 167833 | 2109 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_0179 | Dgeo_0179 | 167919 | 169658 | 1740 | + | PROTEIN:hypothetical protein |
| Dgeo_0180 | Dgeo_0180 | 169746 | 170645 | 900 | - | PROTEIN:4-diphosphocytidyl-2C-methyl-D-erythritol |
| Dgeo_0181 | Dgeo_0181 | 170635 | 171318 | 684 | - | PROTEIN:2-C-methyl-D-erythritol 4-phosphate cytidy |
| Dgeo_0182 | Dgeo_0182 | 171315 | 171911 | 597 | - | PROTEIN:3-octaprenyl-4-hydroxybenzoate carboxy-lya |
| Dgeo_0183 | Dgeo_0183 | 171911 | 172513 | 603 | - | PROTEIN:two component LuxR family transcriptional |
| Dgeo_0184 | Dgeo_0184 | 172510 | 173682 | 1173 | - | PROTEIN:putative signal transduction histidine kin |
| Dgeo_0185 | Dgeo_0185 | 173740 | 174537 | 798 | - | PROTEIN:ABC-2 type transporter |
| Dgeo_0186 | Dgeo_0186 | 174550 | 175089 | 540 | - | PROTEIN:hypothetical protein |
| Dgeo_0187 | Dgeo_0187 | 175086 | 175985 | 900 | - | PROTEIN:ABC transporter related |
| Dgeo_0188 | Dgeo_0188 | 176130 | 176735 | 606 | + | PROTEIN:hypothetical protein |
| Dgeo_0189 | Dgeo_0189 | 176740 | 177501 | 762 | + | PROTEIN:metallophosphoesterase |
| Dgeo_R0003 | Dgeo_R0003 | 177593 | 177668 | 76 | + | RNA:Asp tRNA |
| Dgeo_R0004 | Dgeo_R0004 | 177691 | 177766 | 76 | + | RNA:Phe tRNA |
| Dgeo_0190 | Dgeo_0190 | 177855 | 178580 | 726 | - | PROTEIN:IclR family transcriptional regulator |
| Dgeo_0191 | Dgeo_0191 | 178711 | 178983 | 273 | - | PROTEIN:hypothetical protein |
| Dgeo_0192 | Dgeo_0192 | 178980 | 180077 | 1098 | - | PROTEIN:nucleotidyl transferase |
| Dgeo_0193 | Dgeo_0193 | 180096 | 181013 | 918 | - | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_0194 | Dgeo_0194 | 181095 | 181400 | 306 | + | PROTEIN:hypothetical protein |
| Dgeo_0195 | Dgeo_0195 | 181505 | 182173 | 669 | + | PROTEIN:two component transcriptional regulator |
| Dgeo_0196 | Dgeo_0196 | 182239 | 182727 | 489 | + | PROTEIN:flavin reductase-like protein, FMN-binding |
| Dgeo_0197 | Dgeo_0197 | 182742 | 183011 | 270 | + | PROTEIN:hypothetical protein |
| Dgeo_0198 | Dgeo_0198 | 183060 | 184028 | 969 | + | PROTEIN:cobalamin synthesis protein, P47K |
| Dgeo_0199 | Dgeo_0199 | 184050 | 184517 | 468 | + | PROTEIN:phenylacetic acid degradation-related prot |
| Dgeo_0200 | Dgeo_0200 | 184526 | 185497 | 972 | - | PROTEIN:polysaccharide pyruvyl transferase |
| Dgeo_0201 | Dgeo_0201 | 185494 | 187296 | 1803 | - | PROTEIN:hypothetical protein |
| Dgeo_0202 | Dgeo_0202 | 187525 | 188139 | 615 | - | PROTEIN:uridine kinase |
| Dgeo_0203 | Dgeo_0203 | 188222 | 189256 | 1035 | - | PROTEIN:Glu/Leu/Phe/Val dehydrogenase |
| Dgeo_0204 | Dgeo_0204 | 189505 | 190842 | 1338 | + | PROTEIN:E3 binding |
| Dgeo_0205 | Dgeo_0205 | 191010 | 192224 | 1215 | + | PROTEIN:adenylosuccinate synthetase |
| Dgeo_0206 | folE | 192328 | 192936 | 609 | + | PROTEIN:GTP cyclohydrolase I |
| Dgeo_0207 | Dgeo_0207 | 192980 | 193402 | 423 | - | PROTEIN:SUF system FeS assembly protein |
| Dgeo_0208 | Dgeo_0208 | 193407 | 194600 | 1194 | - | PROTEIN:peptidase M20D, amidohydrolase |
| Dgeo_0209 | Dgeo_0209 | 194651 | 195040 | 390 | + | PROTEIN:hypothetical protein |
| Dgeo_0210 | Dgeo_0210 | 195104 | 196309 | 1206 | + | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_0211 | Dgeo_0211 | 196406 | 197650 | 1245 | - | PROTEIN:VWA containing CoxE-like protein |
| Dgeo_0212 | Dgeo_0212 | 197647 | 198513 | 867 | - | PROTEIN:ATPase |
| Dgeo_0213 | Dgeo_0213 | 198510 | 198974 | 465 | - | PROTEIN:carbon monoxide dehydrogenase subunit G |
| Dgeo_0214 | Dgeo_0214 | 199012 | 199674 | 663 | + | PROTEIN:hypothetical protein |
| Dgeo_0215 | Dgeo_0215 | 199697 | 200596 | 900 | - | PROTEIN:cation diffusion facilitator family transp |
| Dgeo_0216 | Dgeo_0216 | 200593 | 201330 | 738 | - | PROTEIN:CHAD |
| Dgeo_0217 | Dgeo_0217 | 201327 | 202148 | 822 | - | PROTEIN:metallophosphoesterase |
| Dgeo_0218 | Dgeo_0218 | 202145 | 202459 | 315 | - | PROTEIN:hypothetical protein |
| Dgeo_0219 | Dgeo_0219 | 202560 | 203450 | 891 | + | PROTEIN:twin-arginine translocation pathway signal |
| Dgeo_0220 | Dgeo_0220 | 203455 | 204570 | 1116 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_0221 | Dgeo_0221 | 204652 | 205890 | 1239 | - | PROTEIN:glycine hydroxymethyltransferase |
| Dgeo_0222 | cvrA | 205968 | 207467 | 1500 | - | PROTEIN:potassium/proton antiporter |
| Dgeo_0223 | pheS | 207679 | 208698 | 1020 | + | PROTEIN:phenylalanyl-tRNA synthetase subunit alpha |
| Dgeo_0224 | Dgeo_0224 | 208760 | 211219 | 2460 | + | PROTEIN:phenylalanyl-tRNA synthetase, beta subunit |
| Dgeo_0225 | Dgeo_0225 | 211345 | 212619 | 1275 | + | PROTEIN:O-acetylhomoserine aminocarboxypropyltrans |
| Dgeo_0226 | Dgeo_0226 | 212621 | 213226 | 606 | - | PROTEIN:TM2 |
| Dgeo_0227 | Dgeo_0227 | 213312 | 213992 | 681 | + | PROTEIN:isochorismatase hydrolase |
| Dgeo_0228 | Dgeo_0228 | 214016 | 215086 | 1071 | - | PROTEIN:bifunctional nicotinamide mononucleotide a |
| Dgeo_0229 | Dgeo_0229 | 215464 | 216510 | 1047 | + | PROTEIN:protein serine/threonine phosphatases |
| Dgeo_0230 | Dgeo_0230 | 216507 | 216914 | 408 | + | PROTEIN:YjgF-like protein protein |
| Dgeo_0231 | Dgeo_0231 | 217010 | 217849 | 840 | + | PROTEIN:PSP1 |
| Dgeo_0233 | Dgeo_0233 | 218952 | 219245 | 294 | - | PROTEIN:hypothetical protein |
| Dgeo_0234 | Dgeo_0234 | 219242 | 220270 | 1029 | - | PROTEIN:CRISPR-associated Cas1 family protein |
| Dgeo_0235 | Dgeo_0235 | 220267 | 220902 | 636 | - | PROTEIN:CRISPR-associated Cas4 family protein |
| Dgeo_0236 | Dgeo_0236 | 220910 | 221887 | 978 | - | PROTEIN:CRISPR-associated Csd2 family protein |
| Dgeo_0237 | Dgeo_0237 | 221890 | 223875 | 1986 | - | PROTEIN:CRISPR-associated Csd1 family protein |
| Dgeo_0238 | Dgeo_0238 | 223875 | 224630 | 756 | - | PROTEIN:CRISPR-associated Cas5d family protein |
| Dgeo_0239 | Dgeo_0239 | 224788 | 227106 | 2319 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_R0005 | Dgeo_R0005 | 227331 | 227407 | 77 | - | RNA:Pro tRNA |
| Dgeo_0240 | Dgeo_0240 | 227493 | 228743 | 1251 | + | PROTEIN:peptidase M29, aminopeptidase II |
| Dgeo_0241 | mgsA | 228740 | 229153 | 414 | - | PROTEIN:methylglyoxal synthase |
| Dgeo_0242 | Dgeo_0242 | 229150 | 230112 | 963 | - | PROTEIN:ROK domain-containing protein |
| Dgeo_0243 | Dgeo_0243 | 230141 | 230428 | 288 | - | PROTEIN:hypothetical protein |
| Dgeo_0244 | Dgeo_0244 | 230425 | 230916 | 492 | - | PROTEIN:thioesterase superfamily protein |
| Dgeo_0245 | Dgeo_0245 | 230967 | 231362 | 396 | + | PROTEIN:hypothetical protein |
| Dgeo_0246 | Dgeo_0246 | 231412 | 231768 | 357 | + | PROTEIN:CutA1 divalent ion tolerance protein |
| Dgeo_R0006 | Dgeo_R0006 | 231944 | 232019 | 76 | - | RNA:Trp tRNA |
| Dgeo_0247 | Dgeo_0247 | 232111 | 232332 | 222 | - | PROTEIN:hypothetical protein |
| Dgeo_0248 | Dgeo_0248 | 232403 | 233104 | 702 | + | PROTEIN:endonuclease III |
| Dgeo_0249 | Dgeo_0249 | 233172 | 234458 | 1287 | + | PROTEIN:major facilitator transporter |
| Dgeo_0250 | Dgeo_0250 | 234493 | 235209 | 717 | + | PROTEIN:hypothetical protein |
| Dgeo_0251 | Dgeo_0251 | 235507 | 238539 | 3033 | + | PROTEIN:ribonucleoside-diphosphate reductase, aden |
| Dgeo_0252 | Dgeo_0252 | 238873 | 239172 | 300 | + | PROTEIN:DNA-directed RNA polymerase, omega subunit |
| Dgeo_0253 | Dgeo_0253 | 239253 | 240503 | 1251 | + | PROTEIN:sodium/hydrogen exchanger |
| Dgeo_0254 | Dgeo_0254 | 240504 | 242318 | 1815 | - | PROTEIN:cell wall hydrolase/autolysin |
| Dgeo_0255 | Dgeo_0255 | 242553 | 246521 | 3969 | + | PROTEIN:DNA polymerase III, alpha subunit |
| Dgeo_0256 | Dgeo_0256 | 246882 | 247961 | 1080 | - | PROTEIN:twitching motility protein |
| Dgeo_0257 | Dgeo_0257 | 248070 | 248672 | 603 | - | PROTEIN:hypothetical protein |
| Dgeo_0258 | Dgeo_0258 | 248706 | 249626 | 921 | - | PROTEIN:acetyltransferase |
| Dgeo_0259 | Dgeo_0259 | 249780 | 250211 | 432 | - | PROTEIN:isochorismatase hydrolase |
| Dgeo_0260 | Dgeo_0260 | 250238 | 251023 | 786 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0261 | Dgeo_0261 | 251130 | 251897 | 768 | + | PROTEIN:hypothetical protein |
| Dgeo_0262 | Dgeo_0262 | 251894 | 252394 | 501 | + | PROTEIN:HAD family phosphatase |
| Dgeo_0263 | Dgeo_0263 | 252422 | 255097 | 2676 | + | PROTEIN:type II secretion system protein E |
| Dgeo_0264 | Dgeo_0264 | 255238 | 256449 | 1212 | + | PROTEIN:twitching motility protein |
| Dgeo_0265 | Dgeo_0265 | 256472 | 257845 | 1374 | - | PROTEIN:hypothetical protein |
| Dgeo_0266 | Dgeo_0266 | 257975 | 259321 | 1347 | + | PROTEIN:transposase IS66 |
| Dgeo_0267 | Dgeo_0267 | 259414 | 259839 | 426 | + | PROTEIN:hypothetical protein |
| Dgeo_0268 | Dgeo_0268 | 259878 | 261005 | 1128 | + | PROTEIN:nitric-oxide synthase |
| Dgeo_0269 | Dgeo_0269 | 261023 | 263392 | 2370 | - | PROTEIN:penicillin amidase |
| Dgeo_0270 | Dgeo_0270 | 263462 | 264391 | 930 | - | PROTEIN:hypothetical protein |
| Dgeo_0271 | Dgeo_0271 | 264433 | 266958 | 2526 | + | PROTEIN:primosomal protein N' |
| Dgeo_0272 | Dgeo_0272 | 267337 | 268071 | 735 | + | PROTEIN:metallophosphoesterase |
| Dgeo_0273 | Dgeo_0273 | 268112 | 268903 | 792 | - | PROTEIN:hypothetical protein |
| Dgeo_0274 | Dgeo_0274 | 268980 | 270419 | 1440 | - | PROTEIN:hypothetical protein |
| Dgeo_0275 | Dgeo_0275 | 270543 | 270968 | 426 | + | PROTEIN:hypothetical protein |
| Dgeo_0276 | Dgeo_0276 | 271075 | 272160 | 1086 | + | PROTEIN:peptidase M20 |
| Dgeo_0277 | Dgeo_0277 | 272250 | 273593 | 1344 | + | PROTEIN:peptidase S41 |
| Dgeo_0278 | Dgeo_0278 | 273607 | 273978 | 372 | - | PROTEIN:hypothetical protein |
| Dgeo_0279 | Dgeo_0279 | 274131 | 276341 | 2211 | - | PROTEIN:hypothetical protein |
| Dgeo_0280 | Dgeo_0280 | 276550 | 277467 | 918 | + | PROTEIN:beta-lactamase-like protein |
| Dgeo_0281 | Dgeo_0281 | 277636 | 278304 | 669 | + | PROTEIN:Ferritin and Dps |
| Dgeo_0282 | Dgeo_0282 | 278379 | 278870 | 492 | + | PROTEIN:hypothetical protein |
| Dgeo_0283 | Dgeo_0283 | 278963 | 279556 | 594 | - | PROTEIN:molybdopterin-guanine dinucleotide biosynt |
| Dgeo_0284 | Dgeo_0284 | 279751 | 280191 | 441 | + | PROTEIN:hypothetical protein |
| Dgeo_0285 | Dgeo_0285 | 280213 | 280788 | 576 | + | PROTEIN:hypothetical protein |
| Dgeo_0286 | Dgeo_0286 | 280785 | 281339 | 555 | + | PROTEIN:coenzyme A biosynthesis protein |
| Dgeo_0287 | Dgeo_0287 | 281378 | 282604 | 1227 | - | PROTEIN:S-adenosylmethionine synthetase |
| Dgeo_0288 | Dgeo_0288 | 282755 | 283237 | 483 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0289 | Dgeo_0289 | 283418 | 283909 | 492 | - | PROTEIN:hypothetical protein |
| Dgeo_0290 | Dgeo_0290 | 283935 | 284174 | 240 | - | PROTEIN:hypothetical protein |
| Dgeo_0291 | Dgeo_0291 | 284171 | 284530 | 360 | - | PROTEIN:hypothetical protein |
| Dgeo_0292 | Dgeo_0292 | 284614 | 285513 | 900 | - | PROTEIN:arginase |
| Dgeo_0293 | Dgeo_0293 | 285639 | 286319 | 681 | + | PROTEIN:metallophosphoesterase |
| Dgeo_0294 | Dgeo_0294 | 286616 | 287056 | 441 | + | PROTEIN:hypothetical protein |
| Dgeo_0295 | Dgeo_0295 | 287099 | 287635 | 537 | - | PROTEIN:hypothetical protein |
| Dgeo_0296 | Dgeo_0296 | 287677 | 288804 | 1128 | + | PROTEIN:hypothetical protein |
| Dgeo_0297 | Dgeo_0297 | 288792 | 289163 | 372 | - | PROTEIN:phenylacetic acid degradation protein PaaD |
| Dgeo_0298 | Dgeo_0298 | 289216 | 290289 | 1074 | + | PROTEIN:peptidase M42 |
| Dgeo_0299 | Dgeo_0299 | 290273 | 291022 | 750 | + | PROTEIN:silent information regulator protein Sir2 |
| Dgeo_R0007 | Dgeo_R0007 | 291095 | 291247 | 153 | + | RNA:misc_RNA |
| Dgeo_0300 | Dgeo_0300 | 291284 | 292435 | 1152 | + | PROTEIN:riboflavin biosynthesis protein RibD |
| Dgeo_0301 | Dgeo_0301 | 292438 | 293082 | 645 | + | PROTEIN:riboflavin synthase subunit alpha |
| Dgeo_0302 | Dgeo_0302 | 293079 | 294290 | 1212 | + | PROTEIN:GTP cyclohydrolase II |
| Dgeo_0303 | Dgeo_0303 | 294287 | 294751 | 465 | + | PROTEIN:riboflavin synthase |
| Dgeo_0304 | Dgeo_0304 | 294868 | 296358 | 1491 | + | PROTEIN:hypothetical protein |
| Dgeo_0305 | Dgeo_0305 | 296434 | 296829 | 396 | + | PROTEIN:large conductance mechanosensitive channel |
| Dgeo_0306 | Dgeo_0306 | 297150 | 298364 | 1215 | - | PROTEIN:von Willebrand factor, type A |
| Dgeo_0307 | Dgeo_0307 | 298816 | 299223 | 408 | + | PROTEIN:hypothetical protein |
| Dgeo_0308 | Dgeo_0308 | 299257 | 300348 | 1092 | + | PROTEIN:transposase, IS4 |
| Dgeo_0309 | Dgeo_0309 | 300522 | 302810 | 2289 | + | PROTEIN:hypothetical protein |
| Dgeo_0310 | Dgeo_0310 | 302927 | 305701 | 2775 | + | PROTEIN:hypothetical protein |
| Dgeo_0311 | Dgeo_0311 | 305682 | 308873 | 3192 | + | PROTEIN:hypothetical protein |
| Dgeo_0312 | Dgeo_0312 | 308870 | 311197 | 2328 | + | PROTEIN:hypothetical protein |
| Dgeo_0313 | Dgeo_0313 | 311246 | 312199 | 954 | + | PROTEIN:hypothetical protein |
| Dgeo_0314 | Dgeo_0314 | 312196 | 312777 | 582 | - | PROTEIN:hypothetical protein |
| Dgeo_0315 | Dgeo_0315 | 312778 | 313422 | 645 | + | PROTEIN:two component LuxR family transcriptional |
| Dgeo_0316 | Dgeo_0316 | 313628 | 314575 | 948 | + | PROTEIN:beta-lactamase |
| Dgeo_0317 | Dgeo_0317 | 314760 | 315617 | 858 | + | PROTEIN:cytochrome c, class I |
| Dgeo_0318 | Dgeo_0318 | 315643 | 316305 | 663 | + | PROTEIN:Rieske (2Fe-2S) region |
| Dgeo_0319 | Dgeo_0319 | 316302 | 317615 | 1314 | + | PROTEIN:cytochrome b/b6-like protein |
| Dgeo_0320 | Dgeo_0320 | 317684 | 317881 | 198 | + | PROTEIN:hypothetical protein |
| Dgeo_0321 | Dgeo_0321 | 317922 | 318872 | 951 | - | PROTEIN:mannose-6-phosphate isomerase |
| Dgeo_0322 | Dgeo_0322 | 318950 | 319198 | 249 | - | PROTEIN:hypothetical protein |
| Dgeo_0323 | Dgeo_0323 | 319325 | 319747 | 423 | + | PROTEIN:HesB/YadR/YfhF |
| Dgeo_0324 | Dgeo_0324 | 319926 | 321686 | 1761 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_0325 | Dgeo_0325 | 321797 | 322792 | 996 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0326 | Dgeo_0326 | 322803 | 323744 | 942 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0327 | Dgeo_0327 | 323826 | 324332 | 507 | + | PROTEIN:crossover junction endodeoxyribonuclease R |
| Dgeo_0328 | Dgeo_0328 | 324336 | 324986 | 651 | + | PROTEIN:membrane protein-like protein |
| Dgeo_0329 | Dgeo_0329 | 324989 | 325663 | 675 | - | PROTEIN:molybdopterin converting factor, subunit 1 |
| Dgeo_R0008 | Dgeo_R0008 | 325852 | 328741 | 2890 | + | RNA:23S ribosomal RNA |
| Dgeo_R0009 | Dgeo_R0009 | 328825 | 328941 | 117 | + | RNA:5S ribosomal RNA |
| Dgeo_R0010 | Dgeo_R0010 | 328951 | 329026 | 76 | + | RNA:Gly tRNA |
| Dgeo_0330 | Dgeo_0330 | 329158 | 330324 | 1167 | + | PROTEIN:queuine tRNA-ribosyltransferase |
| Dgeo_0331 | Dgeo_0331 | 330767 | 333607 | 2841 | + | PROTEIN:S-layer-like protein region |
| Dgeo_0332 | Dgeo_0332 | 333675 | 334616 | 942 | + | PROTEIN:putative manganese-dependent inorganic pyr |
| Dgeo_0333 | Dgeo_0333 | 334696 | 335940 | 1245 | + | PROTEIN:FolC bifunctional protein |
| Dgeo_0334 | Dgeo_0334 | 336011 | 337504 | 1494 | + | PROTEIN:thermostable carboxypeptidase 1 |
| Dgeo_0335 | Dgeo_0335 | 337594 | 337845 | 252 | - | PROTEIN:hypothetical protein |
| Dgeo_0336 | Dgeo_0336 | 338091 | 338513 | 423 | + | PROTEIN:XRE family transcriptional regulator |
| Dgeo_0337 | Dgeo_0337 | 338569 | 340074 | 1506 | - | PROTEIN:hypothetical protein |
| Dgeo_0338 | moaC | 340114 | 340608 | 495 | - | PROTEIN:molybdenum cofactor biosynthesis protein C |
| Dgeo_0339 | Dgeo_0339 | 340611 | 341261 | 651 | - | PROTEIN:hypothetical protein |
| Dgeo_0340 | Dgeo_0340 | 341415 | 342482 | 1068 | - | PROTEIN:glucose-1-phosphate thymidyltransferase |
| Dgeo_0341 | Dgeo_0341 | 342806 | 343885 | 1080 | + | PROTEIN:mannose-1-phosphate guanylyltransferase (G |
| Dgeo_0342 | Dgeo_0342 | 343882 | 345546 | 1665 | + | PROTEIN:protein-tyrosine kinase |
| Dgeo_0343 | Dgeo_0343 | 345550 | 346992 | 1443 | + | PROTEIN:undecaprenyl-phosphate galactosephosphotra |
| Dgeo_0344 | Dgeo_0344 | 346997 | 348289 | 1293 | + | PROTEIN:polysaccharide biosynthesis protein |
| Dgeo_0345 | Dgeo_0345 | 348293 | 349132 | 840 | + | PROTEIN:glycosyl transferase family protein |
| Dgeo_0346 | Dgeo_0346 | 349122 | 350375 | 1254 | + | PROTEIN:hypothetical protein |
| Dgeo_0347 | Dgeo_0347 | 350359 | 351381 | 1023 | + | PROTEIN:glycosyl transferase, group 1 |
| Dgeo_0348 | Dgeo_0348 | 351371 | 351886 | 516 | + | PROTEIN:Serine O-acetyltransferase |
| Dgeo_0349 | Dgeo_0349 | 351883 | 353079 | 1197 | + | PROTEIN:glycosyl transferase, group 1 |
| Dgeo_0350 | Dgeo_0350 | 353076 | 354035 | 960 | + | PROTEIN:hypothetical protein |
| Dgeo_0351 | Dgeo_0351 | 353968 | 355167 | 1200 | - | PROTEIN:O-antigen polymerase |
| Dgeo_R0011 | Dgeo_R0011 | 355227 | 355301 | 75 | - | RNA:Glu tRNA |
| Dgeo_0352 | Dgeo_0352 | 355442 | 356869 | 1428 | + | PROTEIN:phosphoglucomutase/phosphomannomutase alph |
| Dgeo_0353 | Dgeo_0353 | 356938 | 358344 | 1407 | - | PROTEIN:dihydrolipoamide dehydrogenase |
| Dgeo_0354 | Dgeo_0354 | 358420 | 359073 | 654 | - | PROTEIN:response regulator receiver/unknown domain |
| Dgeo_0355 | Dgeo_0355 | 359070 | 360704 | 1635 | - | PROTEIN:signal transduction histidine kinase regul |
| Dgeo_0356 | Dgeo_0356 | 360988 | 362316 | 1329 | + | PROTEIN:sodium:dicarboxylate symporter |
| Dgeo_0357 | Dgeo_0357 | 362317 | 363258 | 942 | - | PROTEIN:indigoidine synthase A like protein |
| Dgeo_0358 | Dgeo_0358 | 363341 | 364474 | 1134 | - | PROTEIN:PfkB |
| Dgeo_0359 | Dgeo_0359 | 364565 | 365398 | 834 | - | PROTEIN:Citryl-CoA lyase |
| Dgeo_0360 | Dgeo_0360 | 365511 | 365900 | 390 | - | PROTEIN:glyoxalase/bleomycin resistance protein/di |
| Dgeo_0361 | Dgeo_0361 | 365945 | 366685 | 741 | - | PROTEIN:protein of unknown function RIO1 |
| Dgeo_0362 | Dgeo_0362 | 367216 | 368283 | 1068 | + | PROTEIN:Mg2+ transporter protein, CorA-like protei |
| Dgeo_0363 | Dgeo_0363 | 368353 | 369306 | 954 | - | PROTEIN:putative sulfonate/nitrate/taurine transpo |
| Dgeo_0364 | Dgeo_0364 | 369448 | 370194 | 747 | + | PROTEIN:hypothetical protein |
| Dgeo_0365 | Dgeo_0365 | 370256 | 372250 | 1995 | - | PROTEIN:peptidase S9, prolyl oligopeptidase active |
| Dgeo_0366 | Dgeo_0366 | 372704 | 373807 | 1104 | + | PROTEIN:peptidase M20 |
| Dgeo_0367 | Dgeo_0367 | 373809 | 374108 | 300 | + | PROTEIN:hypothetical protein |
| Dgeo_0368 | Dgeo_0368 | 374154 | 374810 | 657 | - | PROTEIN:LmbE-like protein protein |
| Dgeo_0369 | Dgeo_0369 | 374886 | 375881 | 996 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0370 | Dgeo_0370 | 375878 | 376855 | 978 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0371 | Dgeo_0371 | 377034 | 377759 | 726 | + | PROTEIN:hypothetical protein |
| Dgeo_0372 | Dgeo_0372 | 377827 | 378372 | 546 | + | PROTEIN:hypothetical protein |
| Dgeo_0373 | Dgeo_0373 | 378369 | 378743 | 375 | + | PROTEIN:hypothetical protein |
| Dgeo_0374 | Dgeo_0374 | 378749 | 379891 | 1143 | - | PROTEIN:peptidase A24A-like protein |
| Dgeo_0375 | Dgeo_0375 | 379956 | 381413 | 1458 | - | PROTEIN:hypothetical protein |
| Dgeo_0376 | Dgeo_0376 | 381573 | 382373 | 801 | + | PROTEIN:septum site-determining protein MinD |
| Dgeo_0377 | Dgeo_0377 | 382375 | 382632 | 258 | + | PROTEIN:cell division topological specificity fact |
| Dgeo_0378 | Dgeo_0378 | 382694 | 383569 | 876 | + | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0379 | Dgeo_0379 | 383648 | 384592 | 945 | + | PROTEIN:aldo/keto reductase |
| Dgeo_0380 | Dgeo_0380 | 384666 | 386357 | 1692 | - | PROTEIN:diguanylate cyclase |
| Dgeo_0381 | Dgeo_0381 | 386449 | 387444 | 996 | + | PROTEIN:ABC transporter related |
| Dgeo_0382 | Dgeo_0382 | 387437 | 388309 | 873 | + | PROTEIN:hypothetical protein |
| Dgeo_0383 | Dgeo_0383 | 388290 | 388649 | 360 | - | PROTEIN:hypothetical protein |
| Dgeo_0384 | Dgeo_0384 | 388701 | 389141 | 441 | - | PROTEIN:hypothetical protein |
| Dgeo_0385 | Dgeo_0385 | 389245 | 390357 | 1113 | + | PROTEIN:hypothetical protein |
| Dgeo_0386 | Dgeo_0386 | 390546 | 391049 | 504 | + | PROTEIN:ribonuclease activity regulator protein Rr |
| Dgeo_0387 | Dgeo_0387 | 391046 | 391462 | 417 | + | PROTEIN:hypothetical protein |
| Dgeo_0388 | Dgeo_0388 | 391523 | 392527 | 1005 | + | PROTEIN:4Fe-4S ferredoxin, iron-sulfur binding |
| Dgeo_0389 | Dgeo_0389 | 392567 | 394240 | 1674 | - | PROTEIN:histidine kinase |
| Dgeo_0390 | Dgeo_0390 | 394384 | 397014 | 2631 | + | PROTEIN:DEAD/DEAH box helicase-like protein |
| Dgeo_R0012 | Dgeo_R0012 | 397070 | 397143 | 74 | + | RNA:Gln tRNA |
| Dgeo_0391 | Dgeo_0391 | 397384 | 397974 | 591 | + | PROTEIN:S-layer-like protein array-related protein |
| Dgeo_0392 | Dgeo_0392 | 398080 | 398883 | 804 | - | PROTEIN:hypothetical protein |
| Dgeo_0393 | Dgeo_0393 | 398880 | 399989 | 1110 | - | PROTEIN:mandelate racemase/muconate lactonizing-li |
| Dgeo_R0013 | Dgeo_R0013 | 400326 | 401818 | 1493 | + | RNA:16S ribosomal RNA |
| Dgeo_0394 | Dgeo_0394 | 401932 | 402198 | 267 | + | PROTEIN:acyl-CoA-binding protein |
| Dgeo_0395 | Dgeo_0395 | 402295 | 403164 | 870 | + | PROTEIN:hypothetical protein |
| Dgeo_0396 | Dgeo_0396 | 403148 | 404056 | 909 | + | PROTEIN:dihydropteroate synthase |
| Dgeo_0397 | Dgeo_0397 | 404067 | 404441 | 375 | + | PROTEIN:dihydroneopterin aldolase |
| Dgeo_0398 | Dgeo_0398 | 404438 | 404929 | 492 | + | PROTEIN:2-amino-4-hydroxy-6-hydroxymethyldihydropt |
| Dgeo_0399 | Dgeo_0399 | 404971 | 405438 | 468 | + | PROTEIN:hypothetical protein |
| Dgeo_0400 | Dgeo_0400 | 405552 | 405866 | 315 | - | PROTEIN:hypothetical protein |
| Dgeo_0401 | Dgeo_0401 | 406071 | 408236 | 2166 | - | PROTEIN:polynucleotide phosphorylase/polyadenylase |
| Dgeo_0402 | Dgeo_0402 | 408358 | 410439 | 2082 | + | PROTEIN:formate dehydrogenase |
| Dgeo_0403 | Dgeo_0403 | 410517 | 410873 | 357 | - | PROTEIN:arsenate reductase and related |
| Dgeo_0404 | ruvB | 410942 | 411937 | 996 | - | PROTEIN:Holliday junction DNA helicase B |
| Dgeo_0405 | Dgeo_0405 | 412093 | 412782 | 690 | - | PROTEIN:electron transport protein SCO1/SenC |
| Dgeo_0406 | Dgeo_0406 | 412908 | 415364 | 2457 | - | PROTEIN:cytochrome c oxidase, subunit I |
| Dgeo_0407 | Dgeo_0407 | 415361 | 416668 | 1308 | - | PROTEIN:cytochrome c oxidase, subunit II |
| Dgeo_0408 | Dgeo_0408 | 416869 | 417807 | 939 | - | PROTEIN:protoheme IX farnesyltransferase |
| Dgeo_0409 | Dgeo_0409 | 417804 | 418784 | 981 | - | PROTEIN:cytochrome oxidase assembly |
| Dgeo_0410 | Dgeo_0410 | 419061 | 419540 | 480 | + | PROTEIN:hypothetical protein |
| Dgeo_0411 | Dgeo_0411 | 419639 | 420460 | 822 | + | PROTEIN:3-methyl-2-oxobutanoate hydroxymethyltrans |
| Dgeo_0412 | Dgeo_0412 | 420516 | 421976 | 1461 | - | PROTEIN:magnesium protoporphyrin chelatase, putati |
| Dgeo_0413 | Dgeo_0413 | 422044 | 422391 | 348 | - | PROTEIN:hypothetical protein |
| Dgeo_0414 | gatB | 422473 | 424185 | 1713 | + | PROTEIN:aspartyl/glutamyl-tRNA amidotransferase su |
| Dgeo_0415 | Dgeo_0415 | 424223 | 424864 | 642 | + | PROTEIN:hypothetical protein |
| Dgeo_0416 | Dgeo_0416 | 424914 | 425876 | 963 | + | PROTEIN:PfkB |
| Dgeo_0417 | Dgeo_0417 | 425909 | 426904 | 996 | + | PROTEIN:aminodeoxychorismate lyase |
| Dgeo_0418 | Dgeo_0418 | 426958 | 427149 | 192 | + | PROTEIN:hypothetical protein |
| Dgeo_0419 | Dgeo_0419 | 427146 | 427958 | 813 | - | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_0420 | Dgeo_0420 | 427961 | 429655 | 1695 | - | PROTEIN:adenine deaminase |
| Dgeo_0421 | Dgeo_0421 | 429664 | 430446 | 783 | - | PROTEIN:hypothetical protein |
| Dgeo_0422 | Dgeo_0422 | 430487 | 431554 | 1068 | - | PROTEIN:hypothetical protein |
| Dgeo_0423 | Dgeo_0423 | 431609 | 432619 | 1011 | - | PROTEIN:putative esterase |
| Dgeo_0424 | Dgeo_0424 | 432683 | 434380 | 1698 | - | PROTEIN:AMP-dependent synthetase and ligase |
| Dgeo_0425 | Dgeo_0425 | 434566 | 435264 | 699 | - | PROTEIN:Holliday junction resolvase YqgF |
| Dgeo_R0014 | Dgeo_R0014 | 435353 | 435427 | 75 | - | RNA:Asn tRNA |
| Dgeo_0426 | Dgeo_0426 | 435502 | 436032 | 531 | - | PROTEIN:hypothetical protein |
| Dgeo_0427 | Dgeo_0427 | 436153 | 438615 | 2463 | - | PROTEIN:ATP-dependent protease La |
| Dgeo_0428 | Dgeo_0428 | 438926 | 440068 | 1143 | - | PROTEIN:acyl-CoA dehydrogenase-like protein |
| Dgeo_R0015 | Dgeo_R0015 | 440201 | 440277 | 77 | + | RNA:His tRNA |
| Dgeo_R0016 | Dgeo_R0016 | 440331 | 440416 | 86 | + | RNA:Leu tRNA |
| Dgeo_0429 | Dgeo_0429 | 440797 | 441072 | 276 | - | PROTEIN:hypothetical protein |
| Dgeo_0430 | Dgeo_0430 | 441114 | 441560 | 447 | + | PROTEIN:IS1 transposase |
| Dgeo_0431 | Dgeo_0431 | 443273 | 444148 | 876 | - | PROTEIN:phage integrase |
| Dgeo_0432 | Dgeo_0432 | 444527 | 445876 | 1350 | + | PROTEIN:trigger factor-like protein |
| Dgeo_0433 | Dgeo_0433 | 446117 | 447157 | 1041 | + | PROTEIN:3-oxoacyl-(acyl carrier protein) synthase |
| Dgeo_0434 | Dgeo_0434 | 447154 | 448062 | 909 | + | PROTEIN:malonyl CoA-acyl carrier protein transacyl |
| Dgeo_0435 | Dgeo_0435 | 448153 | 448905 | 753 | + | PROTEIN:3-oxoacyl-(acyl-carrier-protein) reductase |
| Dgeo_0436 | Dgeo_0436 | 448998 | 449228 | 231 | + | PROTEIN:acyl carrier protein |
| Dgeo_0437 | Dgeo_0437 | 449343 | 450587 | 1245 | + | PROTEIN:beta-ketoacyl synthase |
| Dgeo_0438 | Dgeo_0438 | 450682 | 452862 | 2181 | + | PROTEIN:polyphosphate kinase |
| Dgeo_0439 | Dgeo_0439 | 453049 | 454941 | 1893 | + | PROTEIN:SARP family transcriptional regulator |
| Dgeo_0440 | Dgeo_0440 | 454948 | 455250 | 303 | + | PROTEIN:hypothetical protein |
| Dgeo_0441 | Dgeo_0441 | 455263 | 455898 | 636 | - | PROTEIN:pyrrolidone-carboxylate peptidase |
| Dgeo_0442 | Dgeo_0442 | 455966 | 456835 | 870 | + | PROTEIN:formamidopyrimidine-DNA glycosylase |
| Dgeo_0443 | Dgeo_0443 | 456828 | 457484 | 657 | + | PROTEIN:pyridoxamine 5'-phosphate oxidase |
| Dgeo_0444 | Dgeo_0444 | 457517 | 458152 | 636 | + | PROTEIN:hypothetical protein |
| Dgeo_0445 | Dgeo_0445 | 458178 | 459794 | 1617 | + | PROTEIN:band 7 protein |
| Dgeo_0446 | Dgeo_0446 | 459988 | 460428 | 441 | + | PROTEIN:OsmC-like protein protein |
| Dgeo_0447 | Dgeo_0447 | 460439 | 461113 | 675 | + | PROTEIN:hypothetical protein |
| Dgeo_0448 | Dgeo_0448 | 461110 | 461517 | 408 | - | PROTEIN:hypothetical protein |
| Dgeo_0449 | tdh | 461925 | 463010 | 1086 | + | PROTEIN:L-threonine 3-dehydrogenase |
| Dgeo_0450 | Dgeo_0450 | 463146 | 464174 | 1029 | + | PROTEIN:tryptophanyl-tRNA synthetase II |
| Dgeo_0451 | Dgeo_0451 | 464251 | 465366 | 1116 | + | PROTEIN:chaperone DnaJ-like protein |
| Dgeo_0452 | Dgeo_0452 | 465437 | 466504 | 1068 | + | PROTEIN:ribose-phosphate pyrophosphokinase |
| Dgeo_0453 | Dgeo_0453 | 466576 | 468048 | 1473 | - | PROTEIN:hypothetical protein |
| Dgeo_0454 | Dgeo_0454 | 468211 | 469059 | 849 | + | PROTEIN:glyoxalase/bleomycin resistance protein/di |
| Dgeo_0455 | Dgeo_0455 | 469106 | 469432 | 327 | + | PROTEIN:Rieske (2Fe-2S) region |
| Dgeo_0456 | Dgeo_0456 | 469446 | 469616 | 171 | - | PROTEIN:hypothetical protein |
| Dgeo_0457 | Dgeo_0457 | 469749 | 470933 | 1185 | - | PROTEIN:peptidase S8 and S53, subtilisin, kexin, s |
| Dgeo_0458 | Dgeo_0458 | 471001 | 471774 | 774 | + | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0459 | Dgeo_0459 | 471844 | 472938 | 1095 | + | PROTEIN:hypothetical protein |
| Dgeo_0460 | Dgeo_0460 | 473076 | 476624 | 3549 | - | PROTEIN:ribonuclease R |
| Dgeo_0461 | Dgeo_0461 | 477037 | 477840 | 804 | - | PROTEIN:exodeoxyribonuclease III (xth) |
| Dgeo_0462 | Dgeo_0462 | 477922 | 478230 | 309 | + | PROTEIN:hypothetical protein |
| Dgeo_0463 | Dgeo_0463 | 478273 | 478866 | 594 | - | PROTEIN:methyltransferase type 11 |
| Dgeo_0464 | Dgeo_0464 | 478946 | 480037 | 1092 | + | PROTEIN:transposase, IS4 |
| Dgeo_0465 | Dgeo_0465 | 480243 | 480461 | 219 | + | PROTEIN:hypothetical protein |
| Dgeo_0466 | Dgeo_0466 | 480682 | 482019 | 1338 | - | PROTEIN:acetyl-CoA carboxylase, biotin carboxylase |
| Dgeo_0467 | Dgeo_0467 | 482106 | 482639 | 534 | - | PROTEIN:acetyl-CoA carboxylase, biotin carboxyl ca |
| Dgeo_0468 | Dgeo_0468 | 482799 | 483356 | 558 | - | PROTEIN:translation elongation factor P |
| Dgeo_0469 | Dgeo_0469 | 483571 | 484296 | 726 | + | PROTEIN:50S ribosomal protein L25/general stress p |
| Dgeo_0471 | Dgeo_0471 | 484906 | 485517 | 612 | - | PROTEIN:hypothetical protein |
| Dgeo_0472 | Dgeo_0472 | 485591 | 486943 | 1353 | - | PROTEIN:FeS assembly protein SufD |
| Dgeo_0473 | Dgeo_0473 | 487002 | 488405 | 1404 | - | PROTEIN:FeS assembly protein SufB |
| Dgeo_0474 | Dgeo_0474 | 488434 | 489204 | 771 | - | PROTEIN:FeS assembly ATPase SufC |
| Dgeo_0475 | Dgeo_0475 | 489603 | 491429 | 1827 | + | PROTEIN:alpha amylase, catalytic region |
| Dgeo_0476 | Dgeo_0476 | 491634 | 493232 | 1599 | + | PROTEIN:RNA polymerase, sigma 28 subunit |
| Dgeo_0477 | Dgeo_0477 | 493455 | 493733 | 279 | + | PROTEIN:hypothetical protein |
| Dgeo_0478 | trmE | 493774 | 495093 | 1320 | - | PROTEIN:tRNA modification GTPase TrmE |
| Dgeo_0479 | Dgeo_0479 | 495554 | 497449 | 1896 | + | PROTEIN:SARP family transcriptional regulator |
| Dgeo_0480 | Dgeo_0480 | 497535 | 498179 | 645 | - | PROTEIN:hypothetical protein |
| Dgeo_0481 | Dgeo_0481 | 498466 | 499329 | 864 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0482 | Dgeo_0482 | 499417 | 499725 | 309 | + | PROTEIN:hypothetical protein |
| Dgeo_0483 | Dgeo_0483 | 499819 | 500598 | 780 | + | PROTEIN:hypothetical protein |
| Dgeo_0484 | Dgeo_0484 | 500639 | 502015 | 1377 | + | PROTEIN:tRNA (uracil-5-)-methyltransferase Gid |
| Dgeo_0485 | Dgeo_0485 | 502032 | 503432 | 1401 | + | PROTEIN:UDP-N-acetylmuramoylalanine--D-glutamate l |
| Dgeo_0486 | Dgeo_0486 | 503429 | 504541 | 1113 | + | PROTEIN:cell cycle protein |
| Dgeo_R0017 | Dgeo_R0017 | 504686 | 504760 | 75 | + | RNA:Glu tRNA |
| Dgeo_0487 | Dgeo_0487 | 504761 | 505498 | 738 | + | PROTEIN:hypothetical protein |
| Dgeo_0488 | Dgeo_0488 | 505571 | 506470 | 900 | + | PROTEIN:Allergen V5/Tpx-1 related |
| Dgeo_0489 | Dgeo_0489 | 506586 | 508148 | 1563 | - | PROTEIN:carboxyl transferase |
| Dgeo_0490 | Dgeo_0490 | 508252 | 508743 | 492 | - | PROTEIN:hypothetical protein |
| Dgeo_0491 | Dgeo_0491 | 508858 | 509466 | 609 | - | PROTEIN:xanthine phosphoribosyltransferase |
| Dgeo_0492 | Dgeo_0492 | 509466 | 510470 | 1005 | - | PROTEIN:hypothetical protein |
| Dgeo_0493 | Dgeo_0493 | 510562 | 511632 | 1071 | + | PROTEIN:acyltransferase 3 |
| Dgeo_0494 | Dgeo_0494 | 511711 | 513033 | 1323 | - | PROTEIN:Glu/Leu/Phe/Val dehydrogenase |
| Dgeo_0495 | Dgeo_0495 | 513141 | 514385 | 1245 | - | PROTEIN:Glu/Leu/Phe/Val dehydrogenase, dimerisatio |
| Dgeo_0496 | Dgeo_0496 | 514701 | 516428 | 1728 | - | PROTEIN:dimethylallyltransferase |
| Dgeo_0497 | Dgeo_0497 | 516425 | 517144 | 720 | - | PROTEIN:hypothetical protein |
| Dgeo_0498 | Dgeo_0498 | 517141 | 519069 | 1929 | - | PROTEIN:glutamine amidotransferase of anthranilate |
| Dgeo_0499 | Dgeo_0499 | 519181 | 519642 | 462 | + | PROTEIN:BadM/Rrf2 family transcriptional regulator |
| Dgeo_0500 | Dgeo_0500 | 519761 | 520846 | 1086 | + | PROTEIN:dihydroorotate dehydrogenase 2 |
| Dgeo_0501 | Dgeo_0501 | 520956 | 521129 | 174 | + | PROTEIN:hypothetical protein |
| Dgeo_0502 | pyrC | 521151 | 522404 | 1254 | - | PROTEIN:dihydroorotase |
| Dgeo_0503 | pyrB | 522532 | 523470 | 939 | - | PROTEIN:aspartate carbamoyltransferase catalytic s |
| Dgeo_0504 | Dgeo_0504 | 523467 | 524021 | 555 | - | PROTEIN:pyrimidine regulatory protein PyrR |
| Dgeo_0505 | Dgeo_0505 | 524385 | 524861 | 477 | + | PROTEIN:heat shock protein Hsp20 |
| Dgeo_0506 | Dgeo_0506 | 524957 | 527032 | 2076 | + | PROTEIN:ABC transporter related |
| Dgeo_0507 | Dgeo_0507 | 527161 | 527619 | 459 | + | PROTEIN:hypothetical protein |
| Dgeo_0508 | Dgeo_0508 | 527699 | 537430 | 9732 | - | PROTEIN:hypothetical protein |
| Dgeo_0509 | Dgeo_0509 | 537589 | 537852 | 264 | + | PROTEIN:acylphosphatase |
| Dgeo_0510 | Dgeo_0510 | 537856 | 538761 | 906 | - | PROTEIN:hypothetical protein |
| Dgeo_0511 | Dgeo_0511 | 538758 | 539405 | 648 | - | PROTEIN:hypothetical protein |
| Dgeo_0512 | Dgeo_0512 | 539484 | 539891 | 408 | + | PROTEIN:endoribonuclease L-PSP |
| Dgeo_0513 | Dgeo_0513 | 540048 | 541592 | 1545 | + | PROTEIN:phosphoribosylaminoimidazolecarboxamide fo |
| Dgeo_0514 | Dgeo_0514 | 541601 | 542467 | 867 | + | PROTEIN:tetrahydrofolate dehydrogenase/cyclohydrol |
| Dgeo_0515 | Dgeo_0515 | 542526 | 543839 | 1314 | - | PROTEIN:glycosyl transferase family protein |
| Dgeo_0516 | Dgeo_0516 | 543938 | 544717 | 780 | - | PROTEIN:major intrinsic protein |
| Dgeo_0517 | Dgeo_0517 | 544759 | 545769 | 1011 | - | PROTEIN:homoserine O-acetyltransferase |
| Dgeo_0518 | Dgeo_0518 | 545766 | 547097 | 1332 | - | PROTEIN:O-acetylhomoserine/O-acetylserine sulfhydr |
| Dgeo_R0018 | Dgeo_R0018 | 547213 | 547339 | 127 | - | RNA:misc_RNA |
| Dgeo_0519 | Dgeo_0519 | 547481 | 547888 | 408 | - | PROTEIN:ferric uptake regulator family protein |
| Dgeo_0520 | Dgeo_0520 | 547979 | 548836 | 858 | - | PROTEIN:hypothetical protein |
| Dgeo_0521 | Dgeo_0521 | 548836 | 549648 | 813 | - | PROTEIN:hypothetical protein |
| Dgeo_0522 | Dgeo_0522 | 549686 | 550390 | 705 | + | PROTEIN:methyltransferase type 11 |
| Dgeo_0523 | Dgeo_0523 | 550460 | 551356 | 897 | + | PROTEIN:squalene/phytoene synthase |
| Dgeo_0524 | Dgeo_0524 | 551432 | 553102 | 1671 | + | PROTEIN:amine oxidase |
| Dgeo_0525 | Dgeo_0525 | 553099 | 553968 | 870 | - | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_0526 | Dgeo_0526 | 554058 | 554495 | 438 | + | PROTEIN:OsmC-like protein protein |
| Dgeo_0527 | Dgeo_0527 | 554588 | 554938 | 351 | - | PROTEIN:HxlR family transcriptional regulator |
| Dgeo_0528 | Dgeo_0528 | 555026 | 555646 | 621 | + | PROTEIN:NADH dehydrogenase |
| Dgeo_0529 | Dgeo_0529 | 555710 | 556687 | 978 | + | PROTEIN:zinc-binding alcohol dehydrogenase |
| Dgeo_0530 | Dgeo_0530 | 556750 | 557997 | 1248 | + | PROTEIN:major facilitator transporter |
| Dgeo_0531 | Dgeo_0531 | 557994 | 558392 | 399 | - | PROTEIN:hypothetical protein |
| Dgeo_0532 | Dgeo_0532 | 558519 | 559328 | 810 | - | PROTEIN:hypothetical protein |
| Dgeo_0533 | Dgeo_0533 | 559402 | 560169 | 768 | - | PROTEIN:ABC transporter related |
| Dgeo_0534 | Dgeo_0534 | 560199 | 561080 | 882 | - | PROTEIN:periplasmic solute binding protein |
| Dgeo_0535 | rpmB | 561080 | 561334 | 255 | - | PROTEIN:50S ribosomal protein L28 |
| Dgeo_0536 | Dgeo_0536 | 561416 | 562474 | 1059 | - | PROTEIN:cobalamin synthesis protein, P47K |
| Dgeo_0537 | Dgeo_0537 | 562673 | 564343 | 1671 | + | PROTEIN:trehalose synthase-like protein |
| Dgeo_0538 | Dgeo_0538 | 564452 | 565138 | 687 | - | PROTEIN:hypothetical protein |
| Dgeo_0539 | Dgeo_0539 | 565216 | 568050 | 2835 | - | PROTEIN:malto-oligosyltrehalose synthase |
| Dgeo_0540 | Dgeo_0540 | 568047 | 569858 | 1812 | - | PROTEIN:malto-oligosyltrehalose trehalohydrolase |
| Dgeo_0541 | Dgeo_0541 | 570089 | 572272 | 2184 | - | PROTEIN:glycogen debranching protein GlgX |
| Dgeo_0542 | Dgeo_0542 | 572466 | 573020 | 555 | - | PROTEIN:hypothetical protein |
| Dgeo_0543 | Dgeo_0543 | 573201 | 574175 | 975 | - | PROTEIN:ABC transporter, substrate-binding protein |
| Dgeo_0544 | Dgeo_0544 | 574203 | 574667 | 465 | + | PROTEIN:hypothetical protein |
| Dgeo_0545 | Dgeo_0545 | 574739 | 577864 | 3126 | + | PROTEIN:transcription factor CarD |
| Dgeo_0546 | Dgeo_0546 | 578107 | 580242 | 2136 | - | PROTEIN:DNA topoisomerase (ATP-hydrolyzing) |
| Dgeo_0547 | Dgeo_0547 | 580350 | 582320 | 1971 | + | PROTEIN:von Willebrand factor, type A |
| Dgeo_R0019 | Dgeo_R0019 | 582403 | 582478 | 76 | - | RNA:Ala tRNA |
| Dgeo_0548 | Dgeo_0548 | 582479 | 584122 | 1644 | + | PROTEIN:inosine-5'-monophosphate dehydrogenase |
| Dgeo_0549 | Dgeo_0549 | 584208 | 584849 | 642 | + | PROTEIN:hemolysin III family channel protein |
| Dgeo_0550 | Dgeo_0550 | 584907 | 585521 | 615 | - | PROTEIN:peptidase M50 |
| Dgeo_0551 | Dgeo_0551 | 585564 | 586697 | 1134 | - | PROTEIN:polynucleotide adenylyltransferase region |
| Dgeo_0552 | Dgeo_0552 | 586701 | 587972 | 1272 | - | PROTEIN:peptidase S1 and S6, chymotrypsin/Hap |
| Dgeo_0553 | Dgeo_0553 | 588189 | 589100 | 912 | - | PROTEIN:Hsp33 protein |
| Dgeo_0554 | Dgeo_0554 | 589456 | 591201 | 1746 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_0555 | Dgeo_0555 | 591277 | 591963 | 687 | + | PROTEIN:two component LuxR family transcriptional |
| Dgeo_0556 | Dgeo_0556 | 591991 | 594960 | 2970 | + | PROTEIN:multi-sensor signal transduction histidine |
| Dgeo_0557 | Dgeo_0557 | 595106 | 595993 | 888 | + | PROTEIN:hypothetical protein |
| Dgeo_0558 | Dgeo_0558 | 596013 | 597038 | 1026 | - | PROTEIN:hypothetical protein |
| Dgeo_0559 | Dgeo_0559 | 597119 | 598354 | 1236 | + | PROTEIN:glycosyl transferase, group 1 |
| Dgeo_0560 | Dgeo_0560 | 598320 | 598997 | 678 | - | PROTEIN:glycosyl transferase family protein |
| Dgeo_0561 | Dgeo_0561 | 599048 | 600256 | 1209 | + | PROTEIN:major facilitator transporter |
| Dgeo_0562 | Dgeo_0562 | 600258 | 600968 | 711 | + | PROTEIN:polysaccharide deacetylase |
| Dgeo_0563 | Dgeo_0563 | 600956 | 602197 | 1242 | - | PROTEIN:hypothetical protein |
| Dgeo_0564 | Dgeo_0564 | 602253 | 602660 | 408 | + | PROTEIN:hypothetical protein |
| Dgeo_0565 | Dgeo_0565 | 602736 | 603701 | 966 | + | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_0566 | Dgeo_0566 | 604158 | 605207 | 1050 | - | PROTEIN:diguanylate cyclase |
| Dgeo_0567 | Dgeo_0567 | 605400 | 607859 | 2460 | - | PROTEIN:leucyl-tRNA synthetase |
| Dgeo_0568 | Dgeo_0568 | 608053 | 608496 | 444 | + | PROTEIN:hypothetical protein |
| Dgeo_0569 | Dgeo_0569 | 608538 | 609191 | 654 | - | PROTEIN:hypothetical protein |
| Dgeo_0570 | Dgeo_0570 | 609253 | 610746 | 1494 | - | PROTEIN:prolyl-tRNA synthetase |
| Dgeo_0571 | Dgeo_0571 | 610902 | 611183 | 282 | + | PROTEIN:hypothetical protein |
| Dgeo_0572 | Dgeo_0572 | 611266 | 613218 | 1953 | + | PROTEIN:alpha amylase, catalytic region |
| Dgeo_0573 | rpmI | 613403 | 613606 | 204 | + | PROTEIN:50S ribosomal protein L35 |
| Dgeo_0574 | rplT | 613612 | 613971 | 360 | + | PROTEIN:50S ribosomal protein L20 |
| Dgeo_0575 | Dgeo_0575 | 614053 | 614865 | 813 | + | PROTEIN:ROK domain-containing protein |
| Dgeo_0576 | Dgeo_0576 | 614885 | 616051 | 1167 | + | PROTEIN:amidohydrolase |
| Dgeo_0577 | rpmE | 616109 | 616327 | 219 | + | PROTEIN:50S ribosomal protein L31 |
| Dgeo_0578 | Dgeo_0578 | 616429 | 617088 | 660 | + | PROTEIN:thymidine kinase |
| Dgeo_0579 | Dgeo_0579 | 617243 | 618349 | 1107 | + | PROTEIN:diguanylate cyclase |
| Dgeo_0580 | Dgeo_0580 | 618368 | 619360 | 993 | - | PROTEIN:LacI family transcription regulator |
| Dgeo_0581 | Dgeo_0581 | 619506 | 619991 | 486 | + | PROTEIN:hypothetical protein |
| Dgeo_0582 | Dgeo_0582 | 620002 | 620649 | 648 | + | PROTEIN:electron transport protein SCO1/SenC |
| Dgeo_0583 | Dgeo_0583 | 620646 | 621473 | 828 | + | PROTEIN:membrane protein |
| Dgeo_0584 | Dgeo_0584 | 621484 | 622977 | 1494 | - | PROTEIN:histidinol dehydrogenase |
| Dgeo_0585 | Dgeo_0585 | 623074 | 623727 | 654 | - | PROTEIN:hypothetical protein |
| Dgeo_0586 | Dgeo_0586 | 623739 | 624917 | 1179 | - | PROTEIN:phosphopentomutase |
| Dgeo_0587 | Dgeo_0587 | 625040 | 625810 | 771 | + | PROTEIN:ABC transporter related |
| Dgeo_0588 | Dgeo_0588 | 625828 | 627534 | 1707 | + | PROTEIN:hypothetical protein |
| Dgeo_0589 | Dgeo_0589 | 627620 | 628471 | 852 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0590 | Dgeo_0590 | 628482 | 629429 | 948 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0591 | Dgeo_0591 | 629514 | 630779 | 1266 | - | PROTEIN:extracellular solute-binding protein |
| Dgeo_0592 | Dgeo_0592 | 630974 | 632722 | 1749 | - | PROTEIN:putative transcriptional regulator |
| Dgeo_0593 | Dgeo_0593 | 632761 | 633405 | 645 | - | PROTEIN:hypothetical protein |
| Dgeo_0594 | Dgeo_0594 | 633557 | 634636 | 1080 | - | PROTEIN:pseudouridylate synthase |
| Dgeo_0595 | Dgeo_0595 | 634690 | 635472 | 783 | - | PROTEIN:hypothetical protein |
| Dgeo_0596 | Dgeo_0596 | 635476 | 636027 | 552 | - | PROTEIN:molybdopterin binding domain-containing pr |
| Dgeo_0597 | Dgeo_0597 | 636153 | 637364 | 1212 | + | PROTEIN:hypothetical protein |
| Dgeo_0598 | guaA | 637520 | 639037 | 1518 | + | PROTEIN:bifunctional GMP synthase/glutamine amidot |
| Dgeo_0599 | Dgeo_0599 | 639140 | 640690 | 1551 | - | PROTEIN:2-isopropylmalate synthase |
| Dgeo_0600 | Dgeo_0600 | 640964 | 641974 | 1011 | - | PROTEIN:ketol-acid reductoisomerase |
| Dgeo_0601 | Dgeo_0601 | 642047 | 642655 | 609 | - | PROTEIN:acetolactate synthase, small subunit |
| Dgeo_0602 | Dgeo_0602 | 642652 | 644373 | 1722 | - | PROTEIN:acetolactate synthase, large subunit, bios |
| Dgeo_0603 | Dgeo_0603 | 644810 | 646585 | 1776 | - | PROTEIN:DEAD/DEAH box helicase-like protein |
| Dgeo_0604 | Dgeo_0604 | 647085 | 648110 | 1026 | - | PROTEIN:pseudouridine synthase, RluD |
| Dgeo_0605 | Dgeo_0605 | 648233 | 648493 | 261 | + | PROTEIN:twin arginine-targeting protein translocas |
| Dgeo_0606 | Dgeo_0606 | 648566 | 649150 | 585 | + | PROTEIN:ECF subfamily RNA polymerase sigma-24 fact |
| Dgeo_0607 | Dgeo_0607 | 649168 | 649800 | 633 | + | PROTEIN:hypothetical protein |
| Dgeo_0608 | Dgeo_0608 | 649836 | 650336 | 501 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_0609 | prfA | 650323 | 651435 | 1113 | - | PROTEIN:peptide chain release factor 1 |
| Dgeo_0610 | Dgeo_0610 | 651568 | 652530 | 963 | + | PROTEIN:homoserine dehydrogenase |
| Dgeo_0611 | Dgeo_0611 | 652543 | 654033 | 1491 | - | PROTEIN:alpha amylase, catalytic region |
| Dgeo_0612 | Dgeo_0612 | 654070 | 655371 | 1302 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_R0020 | Dgeo_R0020 | 655453 | 655527 | 75 | + | RNA:Ala tRNA |
| Dgeo_0613 | Dgeo_0613 | 655642 | 655932 | 291 | - | PROTEIN:hypothetical protein |
| Dgeo_0614 | Dgeo_0614 | 655974 | 656420 | 447 | + | PROTEIN:IS1 transposase |
| Dgeo_0615 | Dgeo_0615 | 656547 | 657299 | 753 | + | PROTEIN:hypothetical protein |
| Dgeo_0616 | Dgeo_0616 | 657296 | 657568 | 273 | + | PROTEIN:hypothetical protein |
| Dgeo_0617 | Dgeo_0617 | 658120 | 660468 | 2349 | + | PROTEIN:3-hydroxyacyl-CoA dehydrogenase, NAD-bindi |
| Dgeo_0618 | Dgeo_0618 | 660717 | 661919 | 1203 | + | PROTEIN:Acetyl-CoA C-acyltransferase |
| Dgeo_0619 | Dgeo_0619 | 662017 | 663228 | 1212 | + | PROTEIN:hypothetical protein |
| Dgeo_0620 | Dgeo_0620 | 663313 | 664509 | 1197 | + | PROTEIN:extracellular ligand-binding receptor |
| Dgeo_0621 | Dgeo_0621 | 664574 | 666955 | 2382 | - | PROTEIN:phosphoenolpyruvate synthase |
| Dgeo_0622 | Dgeo_0622 | 667121 | 667933 | 813 | + | PROTEIN:hypothetical protein |
| Dgeo_0623 | Dgeo_0623 | 668007 | 669284 | 1278 | + | PROTEIN:protein of unknown function DUF224, cystei |
| Dgeo_0624 | Dgeo_0624 | 669281 | 670729 | 1449 | + | PROTEIN:FAD linked oxidase-like protein |
| Dgeo_0625 | Dgeo_0625 | 670716 | 671372 | 657 | + | PROTEIN:glycolate oxidase subunit GlcE |
| Dgeo_0626 | Dgeo_0626 | 671399 | 672670 | 1272 | + | PROTEIN:lactate 2-monooxygenase |
| Dgeo_0627 | cpdB | 672682 | 674601 | 1920 | + | PROTEIN:bifunctional 2',3'-cyclic nucleotide 2'-ph |
| Dgeo_0628 | Dgeo_0628 | 674661 | 679034 | 4374 | - | PROTEIN:hypothetical protein |
| Dgeo_0629 | Dgeo_0629 | 679035 | 679676 | 642 | - | PROTEIN:hypothetical protein |
| Dgeo_0630 | Dgeo_0630 | 679673 | 680368 | 696 | - | PROTEIN:redox-sensing transcriptional repressor Re |
| Dgeo_0631 | Dgeo_0631 | 680373 | 681560 | 1188 | - | PROTEIN:sporulation related |
| Dgeo_0632 | Dgeo_0632 | 681557 | 683182 | 1626 | - | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_0633 | Dgeo_0633 | 683400 | 684434 | 1035 | - | PROTEIN:hypothetical protein |
| Dgeo_0634 | Dgeo_0634 | 684528 | 685043 | 516 | + | PROTEIN:hypothetical protein |
| Dgeo_0635 | Dgeo_0635 | 685249 | 689862 | 4614 | - | PROTEIN:DNA-directed RNA polymerase, subunit beta- |
| Dgeo_0636 | rpoB | 689946 | 693404 | 3459 | - | PROTEIN:DNA-directed RNA polymerase subunit beta |
| Dgeo_0637 | Dgeo_0637 | 693696 | 694496 | 801 | + | PROTEIN:tetratricopeptide TPR_4 |
| Dgeo_0638 | Dgeo_0638 | 694567 | 694830 | 264 | - | PROTEIN:cold-shock DNA-binding domain-containing p |
| Dgeo_0639 | rplL | 695020 | 695388 | 369 | - | PROTEIN:50S ribosomal protein L7/L12 |
| Dgeo_0640 | rplJ | 695443 | 695952 | 510 | - | PROTEIN:50S ribosomal protein L10 |
| Dgeo_0641 | rplA | 696135 | 696836 | 702 | - | PROTEIN:50S ribosomal protein L1 |
| Dgeo_0642 | rplK | 696829 | 697263 | 435 | - | PROTEIN:50S ribosomal protein L11 |
| Dgeo_0643 | Dgeo_0643 | 697406 | 697978 | 573 | - | PROTEIN:NusG antitermination factor |
| Dgeo_0644 | secE | 697975 | 698154 | 180 | - | PROTEIN:preprotein translocase subunit SecE |
| Dgeo_0645 | rpmG | 698238 | 698405 | 168 | - | PROTEIN:50S ribosomal protein L33 |
| Dgeo_0646 | Dgeo_0646 | 698605 | 699822 | 1218 | - | PROTEIN:elongation factor Tu |
| Dgeo_R0021 | Dgeo_R0021 | 699950 | 700024 | 75 | - | RNA:Thr tRNA |
| Dgeo_R0022 | Dgeo_R0022 | 700034 | 700106 | 73 | - | RNA:Gly tRNA |
| Dgeo_R0023 | Dgeo_R0023 | 700110 | 700195 | 86 | - | RNA:Tyr tRNA |
| Dgeo_R0024 | Dgeo_R0024 | 700256 | 700331 | 76 | - | RNA:Thr tRNA |
| Dgeo_0647 | Dgeo_0647 | 700683 | 702608 | 1926 | + | PROTEIN:ABC transporter related |
| Dgeo_0648 | Dgeo_0648 | 702605 | 704476 | 1872 | + | PROTEIN:ABC transporter related |
| Dgeo_0649 | Dgeo_0649 | 704589 | 705611 | 1023 | + | PROTEIN:periplasmic phosphate binding protein |
| Dgeo_0650 | Dgeo_0650 | 705759 | 706772 | 1014 | + | PROTEIN:phosphate ABC transporter permease |
| Dgeo_0651 | Dgeo_0651 | 706769 | 707638 | 870 | + | PROTEIN:phosphate ABC transporter permease |
| Dgeo_0652 | Dgeo_0652 | 707742 | 708500 | 759 | + | PROTEIN:phosphate ABC transporter permease |
| Dgeo_0653 | Dgeo_0653 | 708635 | 709318 | 684 | + | PROTEIN:phosphate uptake regulator, PhoU |
| Dgeo_0654 | Dgeo_0654 | 709574 | 711184 | 1611 | - | PROTEIN:peptidase S49 |
| Dgeo_0655 | Dgeo_0655 | 711189 | 712013 | 825 | - | PROTEIN:globin |
| Dgeo_0656 | Dgeo_0656 | 712072 | 712923 | 852 | - | PROTEIN:cytochrome c, class I |
| Dgeo_0657 | Dgeo_0657 | 713038 | 713775 | 738 | + | PROTEIN:periplasmic binding protein |
| Dgeo_0658 | Dgeo_0658 | 713772 | 714359 | 588 | - | PROTEIN:TetR family transcriptional regulator |
| Dgeo_0659 | Dgeo_0659 | 714413 | 716044 | 1632 | - | PROTEIN:TrkA-C |
| Dgeo_0660 | Dgeo_0660 | 716120 | 717574 | 1455 | - | PROTEIN:alpha amylase, catalytic region |
| Dgeo_0661 | Dgeo_0661 | 717742 | 720237 | 2496 | - | PROTEIN:phosphoenolpyruvate carboxylase |
| Dgeo_0662 | Dgeo_0662 | 720241 | 721032 | 792 | - | PROTEIN:metallophosphoesterase |
| Dgeo_0663 | Dgeo_0663 | 721108 | 722262 | 1155 | + | PROTEIN:putative PAS/PAC sensor protein |
| Dgeo_0664 | Dgeo_0664 | 722269 | 722904 | 636 | - | PROTEIN:hypothetical protein |
| Dgeo_0665 | Dgeo_0665 | 722937 | 724136 | 1200 | + | PROTEIN:N-acetylmuramoyl-L-alanine amidase |
| Dgeo_0666 | Dgeo_0666 | 724169 | 724741 | 573 | - | PROTEIN:cobalamin adenosyltransferase |
| Dgeo_0667 | Dgeo_0667 | 724809 | 726320 | 1512 | - | PROTEIN:4-alpha-glucanotransferase |
| Dgeo_0668 | Dgeo_0668 | 726539 | 727009 | 471 | - | PROTEIN:transcription elongation factor GreA |
| Dgeo_0669 | Dgeo_0669 | 727044 | 728120 | 1077 | - | PROTEIN:twitching motility protein |
| Dgeo_0670 | Dgeo_0670 | 728117 | 729007 | 891 | - | PROTEIN:pantoate--beta-alanine ligase |
| Dgeo_0671 | Dgeo_0671 | 729004 | 729372 | 369 | - | PROTEIN:membrane protein of unknown function |
| Dgeo_0672 | Dgeo_0672 | 729409 | 731010 | 1602 | - | PROTEIN:alpha amylase, catalytic region |
| Dgeo_0673 | Dgeo_0673 | 731530 | 732135 | 606 | - | PROTEIN:septum formation inhibitor MinC |
| Dgeo_0674 | Dgeo_0674 | 732303 | 732686 | 384 | + | PROTEIN:RNA binding S1 |
| Dgeo_0675 | Dgeo_0675 | 732788 | 734446 | 1659 | + | PROTEIN:putative FAD-binding dehydrogenase |
| Dgeo_0676 | Dgeo_0676 | 734561 | 735841 | 1281 | + | PROTEIN:peptidase S1 and S6, chymotrypsin/Hap |
| Dgeo_0677 | Dgeo_0677 | 735891 | 736169 | 279 | - | PROTEIN:hypothetical protein |
| Dgeo_0678 | Dgeo_0678 | 736222 | 737061 | 840 | - | PROTEIN:acetylglutamate kinase |
| Dgeo_0679 | Dgeo_0679 | 737086 | 738159 | 1074 | - | PROTEIN:major facilitator transporter |
| Dgeo_0680 | Dgeo_0680 | 738195 | 739439 | 1245 | - | PROTEIN:transposase, IS605 OrfB |
| Dgeo_0681 | Dgeo_0681 | 739725 | 740030 | 306 | - | PROTEIN:hypothetical protein |
| Dgeo_0682 | Dgeo_0682 | 740027 | 740596 | 570 | - | PROTEIN:hypothetical protein |
| Dgeo_0683 | Dgeo_0683 | 740593 | 741057 | 465 | - | PROTEIN:hypothetical protein |
| Dgeo_0684 | Dgeo_0684 | 741161 | 741607 | 447 | + | PROTEIN:response regulator receiver protein |
| Dgeo_0685 | argC | 741621 | 742670 | 1050 | - | PROTEIN:N-acetyl-gamma-glutamyl-phosphate reductas |
| Dgeo_0686 | Dgeo_0686 | 742795 | 744816 | 2022 | - | PROTEIN:glycoside hydrolase family protein |
| Dgeo_0687 | Dgeo_0687 | 744989 | 746212 | 1224 | - | PROTEIN:extracellular solute-binding protein |
| Dgeo_0688 | Dgeo_0688 | 746322 | 747629 | 1308 | - | PROTEIN:adenylosuccinate lyase |
| Dgeo_0689 | Dgeo_0689 | 747817 | 748506 | 690 | - | PROTEIN:HAD family hydrolase |
| Dgeo_0690 | Dgeo_0690 | 748597 | 749394 | 798 | + | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0691 | Dgeo_0691 | 749490 | 749921 | 432 | - | PROTEIN:disulphide bond formation protein DsbB |
| Dgeo_0692 | Dgeo_0692 | 749932 | 750618 | 687 | - | PROTEIN:DSBA oxidoreductase |
| Dgeo_0693 | Dgeo_0693 | 750683 | 751303 | 621 | - | PROTEIN:hypothetical protein |
| Dgeo_0694 | Dgeo_0694 | 751354 | 754368 | 3015 | - | PROTEIN:excinuclease ABC, A subunit |
| Dgeo_0695 | Dgeo_0695 | 754590 | 755702 | 1113 | - | PROTEIN:basic membrane lipoprotein |
| Dgeo_0696 | Dgeo_0696 | 755915 | 757969 | 2055 | + | PROTEIN:DNA ligase, NAD-dependent |
| Dgeo_0697 | Dgeo_0697 | 757973 | 758431 | 459 | + | PROTEIN:hypothetical protein |
| Dgeo_0698 | Dgeo_0698 | 758428 | 758937 | 510 | + | PROTEIN:NusB antitermination factor |
| Dgeo_0699 | Dgeo_0699 | 758934 | 759803 | 870 | + | PROTEIN:methenyltetrahydrofolate cyclohydrolase |
| Dgeo_0700 | Dgeo_0700 | 759817 | 760278 | 462 | + | PROTEIN:acid phosphatase/vanadium-dependent halope |
| Dgeo_0701 | Dgeo_0701 | 760330 | 760713 | 384 | - | PROTEIN:hypothetical protein |
| Dgeo_0702 | Dgeo_0702 | 760860 | 761123 | 264 | + | PROTEIN:hypothetical protein |
| Dgeo_0703 | Dgeo_0703 | 761133 | 762269 | 1137 | + | PROTEIN:basic membrane lipoprotein |
| Dgeo_0704 | ispG | 762454 | 763728 | 1275 | + | PROTEIN:4-hydroxy-3-methylbut-2-en-1-yl diphosphat |
| Dgeo_0705 | Dgeo_0705 | 763822 | 764223 | 402 | + | PROTEIN:hypothetical protein |
| Dgeo_0706 | Dgeo_0706 | 764267 | 765133 | 867 | - | PROTEIN:peptidase M55, D-aminopeptidase |
| Dgeo_0707 | Dgeo_0707 | 765212 | 765631 | 420 | + | PROTEIN:phenylacetic acid degradation-related prot |
| Dgeo_0708 | aat | 765716 | 766336 | 621 | + | PROTEIN:leucyl/phenylalanyl-tRNA--protein transfer |
| Dgeo_0709 | Dgeo_0709 | 766415 | 767743 | 1329 | - | PROTEIN:manganese transporter NRAMP |
| Dgeo_0710 | Dgeo_0710 | 767862 | 769490 | 1629 | + | PROTEIN:D-3-phosphoglycerate dehydrogenase |
| Dgeo_0711 | Dgeo_0711 | 769870 | 771222 | 1353 | - | PROTEIN:FAD-dependent pyridine nucleotide-disulphi |
| Dgeo_0712 | Dgeo_0712 | 771254 | 772300 | 1047 | + | PROTEIN:hypothetical protein |
| Dgeo_0713 | Dgeo_0713 | 772420 | 773613 | 1194 | + | PROTEIN:aminotransferase, class V |
| Dgeo_0714 | Dgeo_0714 | 773683 | 774318 | 636 | - | PROTEIN:signal transduction protein |
| Dgeo_0715 | Dgeo_0715 | 774390 | 774929 | 540 | + | PROTEIN:outer membrane chaperone Skp (OmpH) |
| Dgeo_0716 | Dgeo_0716 | 775009 | 775485 | 477 | + | PROTEIN:outer membrane chaperone Skp (OmpH) |
| Dgeo_0717 | Dgeo_0717 | 775486 | 775929 | 444 | + | PROTEIN:carboxymuconolactone decarboxylase |
| Dgeo_0718 | Dgeo_0718 | 775949 | 776617 | 669 | - | PROTEIN:hypothetical protein |
| Dgeo_0719 | Dgeo_0719 | 776860 | 777621 | 762 | + | PROTEIN:electron transfer flavoprotein beta-subuni |
| Dgeo_0720 | Dgeo_0720 | 777618 | 778568 | 951 | + | PROTEIN:electron transfer flavoprotein, alpha subu |
| Dgeo_0721 | Dgeo_0721 | 778806 | 779444 | 639 | + | PROTEIN:hypothetical protein |
| Dgeo_0722 | Dgeo_0722 | 779458 | 780792 | 1335 | - | PROTEIN:hypothetical protein |
| Dgeo_0723 | Dgeo_0723 | 780789 | 781688 | 900 | - | PROTEIN:hypothetical protein |
| Dgeo_0724 | Dgeo_0724 | 781714 | 783726 | 2013 | - | PROTEIN:methionyl-tRNA synthetase |
| Dgeo_0725 | Dgeo_0725 | 784017 | 786413 | 2397 | + | PROTEIN:glucan 1,4-alpha-glucosidase |
| Dgeo_0726 | Dgeo_0726 | 786449 | 787045 | 597 | + | PROTEIN:Holliday junction DNA helicase RuvA |
| Dgeo_0727 | Dgeo_0727 | 787028 | 788803 | 1776 | - | PROTEIN:hypothetical protein |
| Dgeo_0728 | Dgeo_0728 | 788946 | 790997 | 2052 | + | PROTEIN:serine/threonine protein kinase |
| Dgeo_0729 | Dgeo_0729 | 791001 | 791264 | 264 | + | PROTEIN:glutaredoxin 2 |
| Dgeo_0730 | Dgeo_0730 | 791300 | 792253 | 954 | + | PROTEIN:signal recognition particle-docking protei |
| Dgeo_0731 | Dgeo_0731 | 792394 | 793104 | 711 | - | PROTEIN:ABC transporter related |
| Dgeo_0732 | Dgeo_0732 | 793104 | 793874 | 771 | - | PROTEIN:ABC transporter related |
| Dgeo_0733 | Dgeo_0733 | 793880 | 794821 | 942 | - | PROTEIN:inner-membrane translocator |
| Dgeo_0734 | Dgeo_0734 | 794823 | 795797 | 975 | - | PROTEIN:inner-membrane translocator |
| Dgeo_0735 | Dgeo_0735 | 795880 | 797037 | 1158 | - | PROTEIN:extracellular ligand-binding receptor |
| Dgeo_0736 | Dgeo_0736 | 797229 | 798368 | 1140 | - | PROTEIN:acyl-CoA dehydrogenase-like protein |
| Dgeo_0737 | Dgeo_0737 | 798540 | 799199 | 660 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0738 | Dgeo_0738 | 799267 | 800535 | 1269 | + | PROTEIN:major facilitator transporter |
| Dgeo_0739 | Dgeo_0739 | 800560 | 801666 | 1107 | - | PROTEIN:methicillin resistance protein |
| Dgeo_0740 | Dgeo_0740 | 801701 | 802738 | 1038 | + | PROTEIN:PpiC-type peptidyl-prolyl cis-trans isomer |
| Dgeo_0741 | Dgeo_0741 | 802761 | 803579 | 819 | - | PROTEIN:hypothetical protein |
| Dgeo_0742 | argJ | 803638 | 804810 | 1173 | - | PROTEIN:bifunctional ornithine acetyltransferase/N |
| Dgeo_0743 | Dgeo_0743 | 804975 | 805496 | 522 | + | PROTEIN:hypothetical protein |
| Dgeo_0744 | Dgeo_0744 | 805595 | 806437 | 843 | + | PROTEIN:serine/threonine protein kinase |
| Dgeo_0745 | Dgeo_0745 | 806428 | 807366 | 939 | + | PROTEIN:DNA polymerase III, delta subunit |
| Dgeo_0746 | Dgeo_0746 | 807376 | 807810 | 435 | + | PROTEIN:hypothetical protein |
| Dgeo_0747 | Dgeo_0747 | 807815 | 808822 | 1008 | - | PROTEIN:DSBA oxidoreductase |
| Dgeo_0748 | Dgeo_0748 | 808870 | 810756 | 1887 | - | PROTEIN:Amylo-alpha-1,6-glucosidase |
| Dgeo_0749 | Dgeo_0749 | 810808 | 812352 | 1545 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0750 | Dgeo_0750 | 812349 | 813449 | 1101 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0751 | Dgeo_0751 | 813519 | 814754 | 1236 | - | PROTEIN:extracellular solute-binding protein |
| Dgeo_0752 | Dgeo_0752 | 814751 | 815746 | 996 | - | PROTEIN:LacI family transcription regulator |
| Dgeo_0753 | Dgeo_0753 | 815926 | 816915 | 990 | - | PROTEIN:zinc-binding alcohol dehydrogenase |
| Dgeo_0754 | Dgeo_0754 | 817036 | 819123 | 2088 | + | PROTEIN:3-hydroxyacyl-CoA dehydrogenase, NAD-bindi |
| Dgeo_0755 | Dgeo_0755 | 819277 | 820458 | 1182 | + | PROTEIN:acetyl-CoA acetyltransferase |
| Dgeo_0756 | Dgeo_0756 | 820615 | 821076 | 462 | + | PROTEIN:protein tyrosine phosphatase |
| Dgeo_0757 | Dgeo_0757 | 821220 | 822440 | 1221 | - | PROTEIN:type II secretion system protein |
| Dgeo_0758 | Dgeo_0758 | 822515 | 823141 | 627 | + | PROTEIN:Sua5/YciO/YrdC/YwlC |
| Dgeo_0759 | Dgeo_0759 | 823138 | 823668 | 531 | + | PROTEIN:putative transcriptional regulator |
| Dgeo_0760 | Dgeo_0760 | 823702 | 825153 | 1452 | + | PROTEIN:glutamyl-tRNA(Gln) amidotransferase, A sub |
| Dgeo_0761 | Dgeo_0761 | 825177 | 826115 | 939 | - | PROTEIN:tRNA pseudouridine synthase B |
| Dgeo_0762 | Dgeo_0762 | 826188 | 826613 | 426 | + | PROTEIN:hypothetical protein |
| Dgeo_0763 | Dgeo_0763 | 826610 | 827371 | 762 | + | PROTEIN:pseudouridine synthase, Rsu |
| Dgeo_R0025 | rnpB | 827461 | 827828 | 368 | + | RNA:RNA component of RNaseP |
| Dgeo_0764 | Dgeo_0764 | 827921 | 828274 | 354 | - | PROTEIN:hypothetical protein |
| Dgeo_0765 | Dgeo_0765 | 828519 | 828947 | 429 | + | PROTEIN:cell division protein MraZ |
| Dgeo_0766 | Dgeo_0766 | 828953 | 829879 | 927 | + | PROTEIN:methyltransferase |
| Dgeo_0767 | Dgeo_0767 | 829876 | 830235 | 360 | + | PROTEIN:hypothetical protein |
| Dgeo_0768 | Dgeo_0768 | 830232 | 831578 | 1347 | + | PROTEIN:peptidoglycan glycosyltransferase |
| Dgeo_0769 | Dgeo_0769 | 831687 | 831932 | 246 | + | PROTEIN:hypothetical protein |
| Dgeo_0770 | Dgeo_0770 | 831997 | 832662 | 666 | + | PROTEIN:hypothetical protein |
| Dgeo_0771 | Dgeo_0771 | 832810 | 833916 | 1107 | - | PROTEIN:alanine dehydrogenase |
| Dgeo_0772 | Dgeo_0772 | 834077 | 834547 | 471 | + | PROTEIN:AsnC family transcriptional regulator |
| Dgeo_0773 | Dgeo_0773 | 834573 | 835259 | 687 | + | PROTEIN:peptidylprolyl isomerase |
| Dgeo_0774 | Dgeo_0774 | 835266 | 835922 | 657 | - | PROTEIN:LrgB-like protein protein |
| Dgeo_0775 | Dgeo_0775 | 835976 | 836362 | 387 | - | PROTEIN:LrgA |
| Dgeo_0776 | Dgeo_0776 | 836359 | 837060 | 702 | - | PROTEIN:adenosylhomocysteine nucleosidase |
| Dgeo_0777 | Dgeo_0777 | 837206 | 837442 | 237 | + | PROTEIN:4Fe-4S ferredoxin, iron-sulfur binding |
| Dgeo_0778 | Dgeo_0778 | 837439 | 837849 | 411 | + | PROTEIN:hypothetical protein |
| Dgeo_0779 | Dgeo_0779 | 837846 | 838253 | 408 | + | PROTEIN:globin |
| Dgeo_0780 | Dgeo_0780 | 838427 | 839122 | 696 | + | PROTEIN:Crp/FNR family transcriptional regulator |
| Dgeo_0781 | Dgeo_0781 | 839279 | 840523 | 1245 | + | PROTEIN:aminotransferase, class I and II |
| Dgeo_0782 | Dgeo_0782 | 840546 | 841208 | 663 | - | PROTEIN:ATP phosphoribosyltransferase catalytic su |
| Dgeo_0783 | hisZ | 841205 | 842392 | 1188 | - | PROTEIN:ATP phosphoribosyltransferase regulatory s |
| Dgeo_0784 | Dgeo_0784 | 842460 | 843032 | 573 | + | PROTEIN:hypothetical protein |
| Dgeo_0785 | Dgeo_0785 | 843038 | 843847 | 810 | + | PROTEIN:HhH-GPD |
| Dgeo_0786 | Dgeo_0786 | 843913 | 844272 | 360 | + | PROTEIN:t-RNA-binding region |
| Dgeo_0787 | Dgeo_0787 | 844406 | 845176 | 771 | - | PROTEIN:integral membrane protein TerC |
| Dgeo_0788 | Dgeo_0788 | 845421 | 846086 | 666 | - | PROTEIN:TetR family transcriptional regulator |
| Dgeo_0789 | Dgeo_0789 | 846094 | 846603 | 510 | + | PROTEIN:hypothetical protein |
| Dgeo_0790 | Dgeo_0790 | 846628 | 847785 | 1158 | - | PROTEIN:diaminopimelate decarboxylase |
| Dgeo_0791 | Dgeo_0791 | 847865 | 848614 | 750 | + | PROTEIN:hypothetical protein |
| Dgeo_0792 | Dgeo_0792 | 848745 | 849890 | 1146 | + | PROTEIN:tRNA (5-methylaminomethyl-2-thiouridylate) |
| Dgeo_0793 | Dgeo_0793 | 849965 | 850855 | 891 | - | PROTEIN:D-isomer specific 2-hydroxyacid dehydrogen |
| Dgeo_0794 | Dgeo_0794 | 850836 | 851402 | 567 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_0795 | Dgeo_0795 | 851413 | 852621 | 1209 | - | PROTEIN:inositol monophosphatase |
| Dgeo_0796 | aspC | 852661 | 853980 | 1320 | + | PROTEIN:aspartyl-tRNA synthetase |
| Dgeo_0797 | Dgeo_0797 | 854023 | 855180 | 1158 | + | PROTEIN:diguanylate cyclase |
| Dgeo_0798 | Dgeo_0798 | 855238 | 855978 | 741 | + | PROTEIN:hydrolase, putative |
| Dgeo_0799 | Dgeo_0799 | 856027 | 856749 | 723 | - | PROTEIN:ABC transporter related |
| Dgeo_0800 | Dgeo_0800 | 856868 | 857629 | 762 | - | PROTEIN:ABC transporter related |
| Dgeo_0801 | Dgeo_0801 | 857626 | 858606 | 981 | - | PROTEIN:inner-membrane translocator |
| Dgeo_0802 | Dgeo_0802 | 858615 | 859475 | 861 | - | PROTEIN:inner-membrane translocator |
| Dgeo_0803 | Dgeo_0803 | 859476 | 860537 | 1062 | - | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_0804 | Dgeo_0804 | 860663 | 860998 | 336 | - | PROTEIN:hypothetical protein |
| Dgeo_0805 | Dgeo_0805 | 861040 | 861486 | 447 | + | PROTEIN:IS1 transposase |
| Dgeo_0806 | Dgeo_0806 | 861478 | 861966 | 489 | - | PROTEIN:leucine-, isoleucine-, valine-, threonine- |
| Dgeo_0807 | Dgeo_0807 | 862073 | 863152 | 1080 | + | PROTEIN:homoserine O-acetyltransferase |
| Dgeo_0808 | Dgeo_0808 | 863399 | 864133 | 735 | + | PROTEIN:3-oxoacid CoA-transferase, subunit A |
| Dgeo_0809 | Dgeo_0809 | 864206 | 864835 | 630 | + | PROTEIN:3-oxoacid CoA-transferase, subunit B |
| Dgeo_0810 | Dgeo_0810 | 865034 | 865510 | 477 | - | PROTEIN:cytochrome c, class I |
| Dgeo_0811 | Dgeo_0811 | 865641 | 867308 | 1668 | + | PROTEIN:propionyl-CoA carboxylase |
| Dgeo_0812 | Dgeo_0812 | 867399 | 868580 | 1182 | + | PROTEIN:acyl-CoA dehydrogenase-like protein |
| Dgeo_0813 | Dgeo_0813 | 869052 | 870224 | 1173 | + | PROTEIN:hypothetical protein |
| Dgeo_0814 | Dgeo_0814 | 870373 | 871398 | 1026 | + | PROTEIN:PEGA domain-containing protein |
| Dgeo_0815 | Dgeo_0815 | 871452 | 872024 | 573 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_0816 | Dgeo_0816 | 872021 | 872653 | 633 | - | PROTEIN:nucleoside triphosphate pyrophosphohydrola |
| Dgeo_0817 | Dgeo_0817 | 872740 | 873216 | 477 | + | PROTEIN:hypothetical protein |
| Dgeo_0818 | Dgeo_0818 | 873279 | 874007 | 729 | + | PROTEIN:short chain dehydrogenase |
| Dgeo_0819 | smpB | 874209 | 874640 | 432 | + | PROTEIN:SsrA-binding protein |
| Dgeo_0820 | Dgeo_0820 | 874637 | 876022 | 1386 | + | PROTEIN:cell wall hydrolase/autolysin |
| Dgeo_0821 | Dgeo_0821 | 876019 | 876558 | 540 | + | PROTEIN:hypothetical protein |
| Dgeo_0822 | Dgeo_0822 | 876600 | 879902 | 3303 | + | PROTEIN:SMC protein-like protein |
| Dgeo_0823 | Dgeo_0823 | 879944 | 882676 | 2733 | - | PROTEIN:SMC protein-like protein |
| Dgeo_0824 | Dgeo_0824 | 882726 | 883913 | 1188 | - | PROTEIN:nuclease SbcCD, D subunit |
| Dgeo_0826 | Dgeo_0826 | 884570 | 886750 | 2181 | + | PROTEIN:helicase RecD/TraA |
| Dgeo_0827 | Dgeo_0827 | 886901 | 888562 | 1662 | + | PROTEIN:CTP synthetase |
| Dgeo_0828 | Dgeo_0828 | 888596 | 889255 | 660 | - | PROTEIN:dephospho-CoA kinase |
| Dgeo_0829 | Dgeo_0829 | 889532 | 890716 | 1185 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_0830 | Dgeo_0830 | 890936 | 891574 | 639 | + | PROTEIN:superoxide dismutase |
| Dgeo_0831 | Dgeo_0831 | 891669 | 893183 | 1515 | - | PROTEIN:hypothetical protein |
| Dgeo_0832 | Dgeo_0832 | 893184 | 893702 | 519 | - | PROTEIN:integral membrane protein |
| Dgeo_0833 | Dgeo_0833 | 893818 | 894765 | 948 | - | PROTEIN:hypothetical protein |
| Dgeo_0834 | Dgeo_0834 | 894966 | 896588 | 1623 | + | PROTEIN:signal transduction histidine kinase regul |
| Dgeo_0835 | Dgeo_0835 | 896585 | 897286 | 702 | + | PROTEIN:response regulator receiver/unknown domain |
| Dgeo_0836 | Dgeo_0836 | 897306 | 897704 | 399 | - | PROTEIN:hypothetical protein |
| Dgeo_0837 | Dgeo_0837 | 897701 | 898084 | 384 | - | PROTEIN:hypothetical protein |
| Dgeo_0838 | Dgeo_0838 | 898140 | 898832 | 693 | - | PROTEIN:hypothetical protein |
| Dgeo_0839 | prmA | 898840 | 899679 | 840 | - | PROTEIN:ribosomal protein L11 methyltransferase |
| Dgeo_0840 | Dgeo_0840 | 899748 | 900257 | 510 | + | PROTEIN:hypothetical protein |
| Dgeo_0841 | Dgeo_0841 | 900302 | 901093 | 792 | + | PROTEIN:pyrroline-5-carboxylate reductase |
| Dgeo_0842 | Dgeo_0842 | 901140 | 901928 | 789 | + | PROTEIN:hypothetical protein |
| Dgeo_0843 | Dgeo_0843 | 901944 | 902573 | 630 | + | PROTEIN:peptide methionine sulfoxide reductase |
| Dgeo_0844 | Dgeo_0844 | 902696 | 903703 | 1008 | - | PROTEIN:transposase, IS4 |
| Dgeo_0845 | Dgeo_0845 | 903796 | 904353 | 558 | + | PROTEIN:hypothetical protein |
| Dgeo_0846 | Dgeo_0846 | 904356 | 905000 | 645 | - | PROTEIN:MOSC domain-containing protein |
| Dgeo_0847 | Dgeo_0847 | 905384 | 906244 | 861 | + | PROTEIN:phosphoribosyltransferase |
| Dgeo_0848 | ubiA | 906549 | 907427 | 879 | + | PROTEIN:prenyltransferase |
| Dgeo_0849 | Dgeo_0849 | 907411 | 908847 | 1437 | + | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_0850 | Dgeo_0850 | 908882 | 910453 | 1572 | - | PROTEIN:1-pyrroline-5-carboxylate dehydrogenase |
| Dgeo_0851 | Dgeo_0851 | 910513 | 911445 | 933 | - | PROTEIN:proline dehydrogenase |
| Dgeo_0852 | Dgeo_0852 | 911442 | 911966 | 525 | - | PROTEIN:GntR family transcriptional regulator |
| Dgeo_0853 | Dgeo_0853 | 911963 | 912418 | 456 | - | PROTEIN:hypothetical protein |
| Dgeo_0854 | Dgeo_0854 | 912468 | 912938 | 471 | - | PROTEIN:hypothetical protein |
| Dgeo_0855 | Dgeo_0855 | 912990 | 913721 | 732 | + | PROTEIN:DNA repair protein RecO |
| Dgeo_0856 | Dgeo_0856 | 913743 | 914627 | 885 | - | PROTEIN:Alpha/beta hydrolase fold-3 |
| Dgeo_0857 | Dgeo_0857 | 914695 | 916002 | 1308 | + | PROTEIN:lycopene cyclase, beta and epsilon |
| Dgeo_0858 | Dgeo_0858 | 915999 | 916295 | 297 | + | PROTEIN:hypothetical protein |
| Dgeo_0859 | Dgeo_0859 | 916292 | 916696 | 405 | - | PROTEIN:heat shock protein Hsp20 |
| Dgeo_0860 | Dgeo_0860 | 916791 | 917705 | 915 | + | PROTEIN:tRNA delta(2)-isopentenylpyrophosphate tra |
| Dgeo_0861 | Dgeo_0861 | 917943 | 919184 | 1242 | + | PROTEIN:glucose-1-phosphate adenylyltransferase |
| Dgeo_0862 | Dgeo_0862 | 919494 | 921140 | 1647 | - | PROTEIN:methylmalonyl-CoA mutase, alpha subunit |
| Dgeo_0863 | Dgeo_0863 | 921299 | 921865 | 567 | + | PROTEIN:peptidase C56, PfpI |
| Dgeo_0864 | Dgeo_0864 | 922097 | 923878 | 1782 | + | PROTEIN:GTP-binding protein TypA |
| Dgeo_0865 | Dgeo_0865 | 923962 | 924912 | 951 | + | PROTEIN:hypothetical protein |
| Dgeo_0866 | Dgeo_0866 | 925010 | 925531 | 522 | + | PROTEIN:hypothetical protein |
| Dgeo_0867 | Dgeo_0867 | 925932 | 926819 | 888 | - | PROTEIN:cation diffusion facilitator family transp |
| Dgeo_0868 | Dgeo_0868 | 927025 | 929250 | 2226 | + | PROTEIN:UvrD/REP helicase |
| Dgeo_0869 | Dgeo_0869 | 929297 | 930688 | 1392 | - | PROTEIN:leucine-rich repeat-containing protein |
| Dgeo_0870 | Dgeo_0870 | 930729 | 931445 | 717 | + | PROTEIN:metallophosphoesterase |
| Dgeo_0871 | Dgeo_0871 | 931558 | 932670 | 1113 | - | PROTEIN:hypothetical protein |
| Dgeo_0872 | Dgeo_0872 | 932768 | 933790 | 1023 | - | PROTEIN:ATPase |
| Dgeo_0873 | Dgeo_0873 | 933818 | 934120 | 303 | - | PROTEIN:hypothetical protein |
| Dgeo_0874 | Dgeo_0874 | 934488 | 937457 | 2970 | + | PROTEIN:protein of unknown function DUF224, cystei |
| Dgeo_0875 | Dgeo_0875 | 937617 | 938312 | 696 | - | PROTEIN:hypothetical protein |
| Dgeo_0876 | Dgeo_0876 | 938481 | 941324 | 2844 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_0877 | Dgeo_0877 | 941422 | 942390 | 969 | + | PROTEIN:putative sulfite oxidase subunit YedY |
| Dgeo_0878 | Dgeo_0878 | 942394 | 943059 | 666 | + | PROTEIN:putative sulfite oxidase subunit YedZ |
| Dgeo_0879 | Dgeo_0879 | 943113 | 943475 | 363 | + | PROTEIN:hypothetical protein |
| Dgeo_0880 | Dgeo_0880 | 943472 | 943675 | 204 | + | PROTEIN:hypothetical protein |
| Dgeo_0881 | Dgeo_0881 | 943672 | 943956 | 285 | + | PROTEIN:hypothetical protein |
| Dgeo_0882 | Dgeo_0882 | 944069 | 945052 | 984 | + | PROTEIN:delta-aminolevulinic acid dehydratase |
| Dgeo_0883 | Dgeo_0883 | 945137 | 945676 | 540 | + | PROTEIN:dual specificity protein phosphatase |
| Dgeo_0884 | Dgeo_0884 | 945693 | 946031 | 339 | + | PROTEIN:antibiotic biosynthesis monooxygenase |
| Dgeo_0885 | Dgeo_0885 | 946101 | 946595 | 495 | + | PROTEIN:hypothetical protein |
| Dgeo_0886 | sdhB | 946691 | 947452 | 762 | - | PROTEIN:succinate dehydrogenase iron-sulfur subuni |
| Dgeo_0887 | sdhA | 947471 | 949222 | 1752 | - | PROTEIN:succinate dehydrogenase flavoprotein subun |
| Dgeo_0888 | Dgeo_0888 | 949354 | 949731 | 378 | - | PROTEIN:succinate dehydrogenase, cytochrome b subu |
| Dgeo_0889 | Dgeo_0889 | 949731 | 950087 | 357 | - | PROTEIN:succinate dehydrogenase, cytochrome b subu |
| Dgeo_0890 | Dgeo_0890 | 950347 | 950697 | 351 | + | PROTEIN:hypothetical protein |
| Dgeo_0891 | Dgeo_0891 | 950804 | 951313 | 510 | - | PROTEIN:pyridoxamine 5'-phosphate oxidase-related, |
| Dgeo_0892 | Dgeo_0892 | 951366 | 952640 | 1275 | - | PROTEIN:major facilitator transporter |
| Dgeo_0893 | Dgeo_0893 | 952695 | 952979 | 285 | - | PROTEIN:hypothetical protein |
| Dgeo_0894 | Dgeo_0894 | 952990 | 955215 | 2226 | - | PROTEIN:ferrous iron transport protein B |
| Dgeo_0895 | Dgeo_0895 | 955212 | 955442 | 231 | - | PROTEIN:hypothetical protein |
| Dgeo_0896 | Dgeo_0896 | 955501 | 956256 | 756 | - | PROTEIN:pseudouridine synthase |
| Dgeo_0897 | Dgeo_0897 | 956291 | 956977 | 687 | - | PROTEIN:hypothetical protein |
| Dgeo_0898 | Dgeo_0898 | 957170 | 958099 | 930 | - | PROTEIN:NAD-dependent epimerase/dehydratase |
| Dgeo_0899 | Dgeo_0899 | 958139 | 960508 | 2370 | + | PROTEIN:MutS2 family protein |
| Dgeo_0900 | Dgeo_0900 | 960700 | 961599 | 900 | + | PROTEIN:hypothetical protein |
| Dgeo_0901 | Dgeo_0901 | 961670 | 962800 | 1131 | + | PROTEIN:major facilitator transporter |
| Dgeo_0902 | Dgeo_0902 | 962954 | 963442 | 489 | + | PROTEIN:GreA/GreB family elongation factor |
| Dgeo_0903 | Dgeo_0903 | 963577 | 965115 | 1539 | + | PROTEIN:lysyl-tRNA synthetase |
| Dgeo_0904 | Dgeo_0904 | 965417 | 966643 | 1227 | + | PROTEIN:ROK domain-containing protein |
| Dgeo_0905 | Dgeo_0905 | 966640 | 967878 | 1239 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_0906 | Dgeo_0906 | 967942 | 969093 | 1152 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0907 | Dgeo_0907 | 969090 | 969953 | 864 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_0908 | Dgeo_0908 | 970075 | 971241 | 1167 | + | PROTEIN:histidine kinase |
| Dgeo_0909 | Dgeo_0909 | 971511 | 971843 | 333 | + | PROTEIN:NADH-ubiquinone/plastoquinone oxidoreducta |
| Dgeo_0910 | Dgeo_0910 | 971857 | 972393 | 537 | + | PROTEIN:NADH-quinone oxidoreductase, B subunit |
| Dgeo_0911 | Dgeo_0911 | 972390 | 973031 | 642 | + | PROTEIN:NADH (or F420H2) dehydrogenase, subunit C |
| Dgeo_0912 | Dgeo_0912 | 973063 | 974274 | 1212 | + | PROTEIN:NADH dehydrogenase I, D subunit |
| Dgeo_0913 | Dgeo_0913 | 974372 | 974983 | 612 | + | PROTEIN:NADH-quinone oxidoreductase, E subunit |
| Dgeo_0914 | Dgeo_0914 | 974980 | 976320 | 1341 | + | PROTEIN:NADH-quinone oxidoreductase, F subunit |
| Dgeo_0915 | Dgeo_0915 | 976379 | 978544 | 2166 | + | PROTEIN:NADH-quinone oxidoreductase, chain G |
| Dgeo_0916 | Dgeo_0916 | 978617 | 979786 | 1170 | + | PROTEIN:respiratory-chain NADH dehydrogenase, subu |
| Dgeo_0917 | Dgeo_0917 | 979841 | 980380 | 540 | + | PROTEIN:NADH-quinone oxidoreductase, chain I |
| Dgeo_0918 | Dgeo_0918 | 980463 | 981056 | 594 | + | PROTEIN:NADH-ubiquinone/plastoquinone oxidoreducta |
| Dgeo_0919 | Dgeo_0919 | 981056 | 981358 | 303 | + | PROTEIN:NADH-ubiquinone oxidoreductase, chain 4L |
| Dgeo_0920 | Dgeo_0920 | 981366 | 983285 | 1920 | + | PROTEIN:proton-translocating NADH-quinone oxidored |
| Dgeo_0921 | Dgeo_0921 | 983392 | 984816 | 1425 | + | PROTEIN:proton-translocating NADH-quinone oxidored |
| Dgeo_0922 | Dgeo_0922 | 984817 | 986265 | 1449 | + | PROTEIN:proton-translocating NADH-quinone oxidored |
| Dgeo_0923 | Dgeo_0923 | 986329 | 988881 | 2553 | - | PROTEIN:type III restriction system endonuclease, |
| Dgeo_0924 | Dgeo_0924 | 988874 | 989293 | 420 | - | PROTEIN:type III restriction system endonuclease, |
| Dgeo_0925 | Dgeo_0925 | 989583 | 990674 | 1092 | + | PROTEIN:transposase, IS4 |
| Dgeo_0926 | Dgeo_0926 | 990649 | 991614 | 966 | - | PROTEIN:Eco57I restriction endonuclease |
| Dgeo_0927 | Dgeo_0927 | 991611 | 992252 | 642 | - | PROTEIN:type III restriction system methylase |
| Dgeo_0928 | Dgeo_0928 | 992283 | 993635 | 1353 | - | PROTEIN:transposase, IS605 OrfB |
| Dgeo_0929 | Dgeo_0929 | 993706 | 995850 | 2145 | + | PROTEIN:ABC transporter related |
| Dgeo_0931 | Dgeo_0931 | 997983 | 999203 | 1221 | - | PROTEIN:toxic anion resistance |
| Dgeo_0932 | Dgeo_0932 | 999267 | 999836 | 570 | - | PROTEIN:hypothetical protein |
| Dgeo_0933 | gatC | 999906 | 1000196 | 291 | + | PROTEIN:aspartyl/glutamyl-tRNA amidotransferase su |
| Dgeo_0934 | Dgeo_0934 | 1000245 | 1001558 | 1314 | + | PROTEIN:seryl-tRNA synthetase |
| Dgeo_0935 | Dgeo_0935 | 1001693 | 1002097 | 405 | + | PROTEIN:hypothetical protein |
| Dgeo_0936 | Dgeo_0936 | 1002116 | 1002409 | 294 | - | PROTEIN:hypothetical protein |
| Dgeo_0937 | Dgeo_0937 | 1002590 | 1003090 | 501 | + | PROTEIN:hypothetical protein |
| Dgeo_0938 | Dgeo_0938 | 1003092 | 1003715 | 624 | + | PROTEIN:competence protein ComF, putative |
| Dgeo_0939 | Dgeo_0939 | 1003767 | 1004288 | 522 | + | PROTEIN:hypothetical protein |
| Dgeo_0940 | Dgeo_0940 | 1004331 | 1005026 | 696 | + | PROTEIN:hypothetical protein |
| Dgeo_0941 | Dgeo_0941 | 1005063 | 1006721 | 1659 | - | PROTEIN:phospholipase/carboxylesterase |
| Dgeo_0942 | Dgeo_0942 | 1006792 | 1007286 | 495 | - | PROTEIN:MarR family transcriptional regulator |
| Dgeo_0943 | Dgeo_0943 | 1007364 | 1008632 | 1269 | - | PROTEIN:hypothetical protein |
| Dgeo_0944 | Dgeo_0944 | 1008706 | 1009896 | 1191 | + | PROTEIN:cytochrome P450 |
| Dgeo_0945 | Dgeo_0945 | 1010040 | 1010585 | 546 | + | PROTEIN:hypothetical protein |
| Dgeo_0946 | Dgeo_0946 | 1010596 | 1011696 | 1101 | + | PROTEIN:queuosine biosynthesis protein |
| Dgeo_0947 | Dgeo_0947 | 1011734 | 1012276 | 543 | + | PROTEIN:bifunctional deaminase-reductase-like prot |
| Dgeo_0948 | Dgeo_0948 | 1012473 | 1014545 | 2073 | + | PROTEIN:oligopeptidase A |
| Dgeo_0949 | Dgeo_0949 | 1014639 | 1016360 | 1722 | + | PROTEIN:hypothetical protein |
| Dgeo_0950 | Dgeo_0950 | 1016464 | 1017546 | 1083 | + | PROTEIN:hypothetical protein |
| Dgeo_0951 | Dgeo_0951 | 1017543 | 1018772 | 1230 | + | PROTEIN:polysaccharide deacetylase |
| Dgeo_0952 | Dgeo_0952 | 1018784 | 1019560 | 777 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0953 | Dgeo_0953 | 1019618 | 1020589 | 972 | - | PROTEIN:beta-lactamase |
| Dgeo_0954 | Dgeo_0954 | 1020586 | 1023567 | 2982 | - | PROTEIN:TPR repeat-containing protein |
| Dgeo_0955 | Dgeo_0955 | 1023588 | 1024103 | 516 | - | PROTEIN:chromate transporter |
| Dgeo_0956 | Dgeo_0956 | 1024096 | 1024686 | 591 | - | PROTEIN:chromate transporter |
| Dgeo_0958 | Dgeo_0958 | 1025204 | 1026244 | 1041 | - | PROTEIN:CRISPR-associated APE2256 family protein |
| Dgeo_0959 | Dgeo_0959 | 1026241 | 1027428 | 1188 | - | PROTEIN:CRISPR-associated TM1812 family protein |
| Dgeo_0960 | Dgeo_0960 | 1027425 | 1028453 | 1029 | - | PROTEIN:CRISPR-associated Cmr5 family protein |
| Dgeo_0961 | Dgeo_0961 | 1028450 | 1028962 | 513 | - | PROTEIN:CRISPR-associated Cmr5 family protein |
| Dgeo_0962 | Dgeo_0962 | 1028959 | 1029753 | 795 | - | PROTEIN:CRISPR-associated RAMP Cmr4 family protein |
| Dgeo_0963 | Dgeo_0963 | 1029753 | 1030937 | 1185 | - | PROTEIN:hypothetical protein |
| Dgeo_0964 | Dgeo_0964 | 1030934 | 1032367 | 1434 | - | PROTEIN:CRISPR-associated Cmr2 family protein |
| Dgeo_0965 | Dgeo_0965 | 1032364 | 1033572 | 1209 | - | PROTEIN:CRISPR-associated Cmr1 family protein |
| Dgeo_0966 | Dgeo_0966 | 1033836 | 1034945 | 1110 | + | PROTEIN:NADH:flavin oxidoreductase/NADH oxidase |
| Dgeo_0967 | Dgeo_0967 | 1034976 | 1035761 | 786 | - | PROTEIN:ABC transporter related |
| Dgeo_0968 | Dgeo_0968 | 1035828 | 1036460 | 633 | + | PROTEIN:Crp/FNR family transcriptional regulator |
| Dgeo_0969 | Dgeo_0969 | 1036548 | 1037735 | 1188 | - | PROTEIN:hypothetical protein |
| Dgeo_0970 | Dgeo_0970 | 1038277 | 1038987 | 711 | + | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_0971 | Dgeo_0971 | 1038984 | 1039529 | 546 | + | PROTEIN:hypothetical protein |
| Dgeo_0972 | Dgeo_0972 | 1039679 | 1043479 | 3801 | + | PROTEIN:endonuclease/exonuclease/phosphatase |
| Dgeo_0973 | Dgeo_0973 | 1043535 | 1043894 | 360 | + | PROTEIN:inorganic pyrophosphatase-related protein |
| Dgeo_0974 | Dgeo_0974 | 1043904 | 1044596 | 693 | + | PROTEIN:short chain dehydrogenase |
| Dgeo_0975 | Dgeo_0975 | 1044604 | 1045566 | 963 | + | PROTEIN:FAD-dependent pyridine nucleotide-disulphi |
| Dgeo_0976 | Dgeo_0976 | 1045563 | 1048109 | 2547 | + | PROTEIN:ATP-dependent helicase HrpB |
| Dgeo_0977 | Dgeo_0977 | 1048268 | 1048873 | 606 | - | PROTEIN:Rad52/22 double-strand break repair protei |
| Dgeo_0978 | Dgeo_0978 | 1049083 | 1050627 | 1545 | - | PROTEIN:peptidase S8 and S53, subtilisin, kexin, s |
| Dgeo_0979 | Dgeo_0979 | 1051007 | 1051618 | 612 | + | PROTEIN:imidazoleglycerol-phosphate dehydratase |
| Dgeo_0980 | hisH | 1051615 | 1052253 | 639 | + | PROTEIN:imidazole glycerol phosphate synthase subu |
| Dgeo_0981 | Dgeo_0981 | 1052368 | 1054326 | 1959 | + | PROTEIN:glycogen branching enzyme |
| Dgeo_0982 | Dgeo_0982 | 1054323 | 1054598 | 276 | + | PROTEIN:hypothetical protein |
| Dgeo_0983 | Dgeo_0983 | 1054984 | 1056408 | 1425 | + | PROTEIN:anthranilate synthase component I |
| Dgeo_0984 | Dgeo_0984 | 1056423 | 1056809 | 387 | + | PROTEIN:S23 ribosomal |
| Dgeo_0985 | Dgeo_0985 | 1056806 | 1057447 | 642 | + | PROTEIN:anthranilate synthase component II |
| Dgeo_0986 | Dgeo_0986 | 1057444 | 1058499 | 1056 | + | PROTEIN:anthranilate phosphoribosyltransferase |
| Dgeo_0987 | Dgeo_0987 | 1058503 | 1058964 | 462 | - | PROTEIN:AsnC family transcriptional regulator |
| Dgeo_0988 | Dgeo_0988 | 1059124 | 1060575 | 1452 | + | PROTEIN:hypothetical protein |
| Dgeo_0989 | Dgeo_0989 | 1060684 | 1062060 | 1377 | + | PROTEIN:4-aminobutyrate aminotransferase |
| Dgeo_0990 | Dgeo_0990 | 1062057 | 1062515 | 459 | - | PROTEIN:redoxin |
| Dgeo_0991 | Dgeo_0991 | 1062593 | 1064212 | 1620 | + | PROTEIN:tRNA(Ile)-lysidine synthetase-like protein |
| Dgeo_0992 | Dgeo_0992 | 1064279 | 1065370 | 1092 | + | PROTEIN:gamma-glutamyl kinase |
| Dgeo_0993 | Dgeo_0993 | 1065381 | 1066730 | 1350 | + | PROTEIN:gamma-glutamyl phosphate reductase |
| Dgeo_0994 | Dgeo_0994 | 1066801 | 1068702 | 1902 | - | PROTEIN:1-deoxy-D-xylulose-5-phosphate synthase |
| Dgeo_0995 | Dgeo_0995 | 1068776 | 1069573 | 798 | - | PROTEIN:hypothetical protein |
| Dgeo_0996 | Dgeo_0996 | 1069615 | 1070328 | 714 | - | PROTEIN:phage shock protein A, PspA |
| Dgeo_0997 | Dgeo_0997 | 1070454 | 1071254 | 801 | - | PROTEIN:hypothetical protein |
| Dgeo_0998 | Dgeo_0998 | 1071327 | 1072412 | 1086 | + | PROTEIN:PilT protein-like protein |
| Dgeo_0999 | Dgeo_0999 | 1072506 | 1073723 | 1218 | - | PROTEIN:saccharopine dehydrogenase |
| Dgeo_1000 | Dgeo_1000 | 1073805 | 1074134 | 330 | + | PROTEIN:hypothetical protein |
| Dgeo_1001 | Dgeo_1001 | 1074199 | 1075521 | 1323 | + | PROTEIN:3-phosphoshikimate 1-carboxyvinyltransfera |
| Dgeo_1002 | Dgeo_1002 | 1075604 | 1076695 | 1092 | + | PROTEIN:NHL repeat-containing protein |
| Dgeo_1003 | Dgeo_1003 | 1076768 | 1077580 | 813 | + | PROTEIN:nitroreductase |
| Dgeo_1004 | Dgeo_1004 | 1077642 | 1078826 | 1185 | + | PROTEIN:cystathionine gamma-synthase |
| Dgeo_1005 | Dgeo_1005 | 1078845 | 1079408 | 564 | + | PROTEIN:hypothetical protein |
| Dgeo_1006 | Dgeo_1006 | 1079568 | 1079837 | 270 | - | PROTEIN:cold-shock DNA-binding domain-containing p |
| Dgeo_1007 | Dgeo_1007 | 1080005 | 1080439 | 435 | + | PROTEIN:NUDIX hydrolase |
| Dgeo_1008 | Dgeo_1008 | 1080460 | 1080990 | 531 | + | PROTEIN:ferripyochelin-binding protein |
| Dgeo_1009 | Dgeo_1009 | 1080987 | 1081817 | 831 | + | PROTEIN:hypothetical protein |
| Dgeo_1010 | Dgeo_1010 | 1081831 | 1082826 | 996 | - | PROTEIN:hydroxymethylbutenyl pyrophosphate reducta |
| Dgeo_1011 | Dgeo_1011 | 1082896 | 1084386 | 1491 | - | PROTEIN:hypothetical protein |
| Dgeo_1012 | Dgeo_1012 | 1084468 | 1086150 | 1683 | - | PROTEIN:hypothetical protein |
| Dgeo_1013 | Dgeo_1013 | 1086200 | 1087195 | 996 | - | PROTEIN:FAD-dependent pyridine nucleotide-disulphi |
| Dgeo_1014 | Dgeo_1014 | 1087266 | 1088660 | 1395 | - | PROTEIN:FAD-dependent pyridine nucleotide-disulphi |
| Dgeo_1015 | Dgeo_1015 | 1088822 | 1089436 | 615 | - | PROTEIN:Crp/FNR family transcriptional regulator |
| Dgeo_1016 | Dgeo_1016 | 1089765 | 1092194 | 2430 | - | PROTEIN:DNA gyrase, A subunit |
| Dgeo_1017 | Dgeo_1017 | 1092231 | 1092533 | 303 | + | PROTEIN:hypothetical protein |
| Dgeo_1018 | Dgeo_1018 | 1092533 | 1092883 | 351 | + | PROTEIN:roadblock/LC7 domain-contain protein |
| Dgeo_1019 | Dgeo_1019 | 1092880 | 1093221 | 342 | + | PROTEIN:hypothetical protein |
| Dgeo_1020 | Dgeo_1020 | 1093218 | 1093517 | 300 | + | PROTEIN:hypothetical protein |
| Dgeo_1021 | Dgeo_1021 | 1093581 | 1094480 | 900 | - | PROTEIN:LmbE-like protein protein |
| Dgeo_1022 | Dgeo_1022 | 1094500 | 1094724 | 225 | - | PROTEIN:hypothetical protein |
| Dgeo_1023 | Dgeo_1023 | 1094755 | 1095666 | 912 | + | PROTEIN:glutamyl-tRNA synthetase, class Ic |
| Dgeo_1024 | Dgeo_1024 | 1095724 | 1096968 | 1245 | + | PROTEIN:tryptophan synthase subunit beta |
| Dgeo_1025 | trpA | 1096965 | 1097765 | 801 | + | PROTEIN:tryptophan synthase subunit alpha |
| Dgeo_1026 | Dgeo_1026 | 1097841 | 1098404 | 564 | - | PROTEIN:hypothetical protein |
| Dgeo_1027 | Dgeo_1027 | 1098468 | 1098950 | 483 | - | PROTEIN:hypothetical protein |
| Dgeo_1029 | Dgeo_1029 | 1099835 | 1100683 | 849 | + | PROTEIN:beta-lactamase-like protein |
| Dgeo_1030 | Dgeo_1030 | 1100915 | 1102210 | 1296 | + | PROTEIN:3-isopropylmalate dehydratase large subuni |
| Dgeo_1031 | Dgeo_1031 | 1102236 | 1103528 | 1293 | + | PROTEIN:hypothetical protein |
| Dgeo_1032 | Dgeo_1032 | 1103560 | 1104084 | 525 | + | PROTEIN:3-isopropylmalate dehydratase, small subun |
| Dgeo_1033 | Dgeo_1033 | 1104077 | 1105132 | 1056 | + | PROTEIN:3-isopropylmalate dehydrogenase |
| Dgeo_1034 | Dgeo_1034 | 1105136 | 1106950 | 1815 | + | PROTEIN:cell wall hydrolase/autolysin |
| Dgeo_1035 | Dgeo_1035 | 1106997 | 1107527 | 531 | + | PROTEIN:hypothetical protein |
| Dgeo_1036 | argS | 1107543 | 1109429 | 1887 | + | PROTEIN:arginyl-tRNA synthetase |
| Dgeo_1037 | Dgeo_1037 | 1109551 | 1110135 | 585 | + | PROTEIN:hypothetical protein |
| Dgeo_1038 | Dgeo_1038 | 1110198 | 1111064 | 867 | - | PROTEIN:hypothetical protein |
| Dgeo_1039 | Dgeo_1039 | 1111079 | 1112212 | 1134 | - | PROTEIN:acyltransferase 3 |
| Dgeo_1040 | Dgeo_1040 | 1112265 | 1113377 | 1113 | - | PROTEIN:prephenate dehydrogenase |
| Dgeo_1041 | Dgeo_1041 | 1113427 | 1114095 | 669 | - | PROTEIN:glycosyl transferase family protein |
| Dgeo_1042 | Dgeo_1042 | 1114171 | 1114968 | 798 | - | PROTEIN:transposase, IS4 |
| Dgeo_1043 | Dgeo_1043 | 1115030 | 1116148 | 1119 | - | PROTEIN:peptidase M50 |
| Dgeo_1044 | Dgeo_1044 | 1116145 | 1117308 | 1164 | - | PROTEIN:1-deoxy-D-xylulose 5-phosphate reductoisom |
| Dgeo_1045 | Dgeo_1045 | 1117384 | 1118214 | 831 | - | PROTEIN:phosphatidate cytidylyltransferase |
| Dgeo_1046 | Dgeo_1046 | 1118327 | 1118878 | 552 | - | PROTEIN:ribosome recycling factor |
| Dgeo_1047 | Dgeo_1047 | 1118902 | 1119675 | 774 | - | PROTEIN:uridylate kinase |
| Dgeo_1048 | tsf | 1119792 | 1120601 | 810 | - | PROTEIN:elongation factor Ts |
| Dgeo_1049 | rpsB | 1120691 | 1121485 | 795 | - | PROTEIN:30S ribosomal protein S2 |
| Dgeo_1050 | Dgeo_1050 | 1121526 | 1121744 | 219 | + | PROTEIN:hypothetical protein |
| Dgeo_1051 | Dgeo_1051 | 1121757 | 1122581 | 825 | + | PROTEIN:undecaprenol kinase, putative |
| Dgeo_1052 | Dgeo_1052 | 1122592 | 1122954 | 363 | + | PROTEIN:hypothetical protein |
| Dgeo_1053 | Dgeo_1053 | 1123021 | 1123263 | 243 | + | PROTEIN:hypothetical protein |
| Dgeo_1054 | Dgeo_1054 | 1123260 | 1123697 | 438 | + | PROTEIN:hypothetical protein |
| Dgeo_1055 | Dgeo_1055 | 1123694 | 1124134 | 441 | + | PROTEIN:guanine-specific ribonuclease N1 and T1 |
| Dgeo_1056 | Dgeo_1056 | 1124131 | 1124529 | 399 | + | PROTEIN:hypothetical protein |
| Dgeo_1057 | Dgeo_1057 | 1124574 | 1125575 | 1002 | + | PROTEIN:O-sialoglycoprotein endopeptidase |
| Dgeo_1058 | secA | 1125658 | 1128267 | 2610 | + | PROTEIN:preprotein translocase subunit SecA |
| Dgeo_1059 | Dgeo_1059 | 1128462 | 1129670 | 1209 | + | PROTEIN:ABC transporter related |
| Dgeo_1060 | Dgeo_1060 | 1129861 | 1130619 | 759 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_1061 | Dgeo_1061 | 1130777 | 1131568 | 792 | + | PROTEIN:amino acid ABC transporter permease |
| Dgeo_1062 | Dgeo_1062 | 1131674 | 1133854 | 2181 | - | PROTEIN:TRAP transporter, 4TM/12TM fusion protein |
| Dgeo_1063 | Dgeo_1063 | 1133937 | 1134890 | 954 | - | PROTEIN:TRAP transporter solute receptor TAXI fami |
| Dgeo_1064 | Dgeo_1064 | 1134985 | 1135743 | 759 | + | PROTEIN:ABC transporter related |
| Dgeo_1065 | Dgeo_1065 | 1135758 | 1137017 | 1260 | - | PROTEIN:major facilitator transporter |
| Dgeo_1066 | Dgeo_1066 | 1137050 | 1137835 | 786 | + | PROTEIN:WecB/TagA/CpsF family glycosyl transferase |
| Dgeo_1067 | Dgeo_1067 | 1137849 | 1138250 | 402 | - | PROTEIN:penicillin-binding protein 1B McrB, putati |
| Dgeo_1068 | Dgeo_1068 | 1138373 | 1140721 | 2349 | - | PROTEIN:peptidoglycan glycosyltransferase |
| Dgeo_1069 | Dgeo_1069 | 1140759 | 1141145 | 387 | - | PROTEIN:response regulator receiver protein |
| Dgeo_1070 | Dgeo_1070 | 1141311 | 1143008 | 1698 | - | PROTEIN:AMP-dependent synthetase and ligase |
| Dgeo_1071 | Dgeo_1071 | 1143119 | 1143682 | 564 | + | PROTEIN:CMP/dCMP deaminase, zinc-binding |
| Dgeo_1072 | Dgeo_1072 | 1143679 | 1144566 | 888 | + | PROTEIN:PfkB |
| Dgeo_1073 | Dgeo_1073 | 1144576 | 1145091 | 516 | + | PROTEIN:hypothetical protein |
| Dgeo_1074 | Dgeo_1074 | 1145091 | 1145315 | 225 | + | PROTEIN:hypothetical protein |
| Dgeo_1075 | Dgeo_1075 | 1145321 | 1145632 | 312 | + | PROTEIN:hypothetical protein |
| Dgeo_1076 | Dgeo_1076 | 1145710 | 1146489 | 780 | - | PROTEIN:peptidase S26A, signal peptidase I |
| Dgeo_1077 | Dgeo_1077 | 1146559 | 1147350 | 792 | - | PROTEIN:patatin |
| Dgeo_1078 | Dgeo_1078 | 1147418 | 1148170 | 753 | + | PROTEIN:serine/threonine protein kinase |
| Dgeo_1079 | Dgeo_1079 | 1148234 | 1149670 | 1437 | + | PROTEIN:Beta-N-acetylhexosaminidase |
| Dgeo_1080 | Dgeo_1080 | 1149788 | 1150669 | 882 | - | PROTEIN:hypothetical protein |
| Dgeo_1081 | Dgeo_1081 | 1150666 | 1151553 | 888 | - | PROTEIN:6-phosphogluconate dehydrogenase, NAD-bind |
| Dgeo_1082 | Dgeo_1082 | 1151635 | 1152543 | 909 | + | PROTEIN:auxin efflux carrier |
| Dgeo_1083 | Dgeo_1083 | 1152594 | 1153385 | 792 | + | PROTEIN:tRNA/rRNA methyltransferase (SpoU) |
| Dgeo_1084 | Dgeo_1084 | 1153437 | 1155254 | 1818 | + | PROTEIN:oligoendopeptidase F |
| Dgeo_1085 | Dgeo_1085 | 1155309 | 1155770 | 462 | - | PROTEIN:hypothetical protein |
| Dgeo_1086 | Dgeo_1086 | 1155829 | 1156320 | 492 | - | PROTEIN:rare lipoprotein A |
| Dgeo_1087 | Dgeo_1087 | 1156317 | 1157363 | 1047 | - | PROTEIN:peptidase M24 |
| Dgeo_1088 | Dgeo_1088 | 1157522 | 1158679 | 1158 | + | PROTEIN:succinyl-CoA synthetase, beta subunit |
| Dgeo_1089 | Dgeo_1089 | 1158679 | 1159590 | 912 | + | PROTEIN:succinyl-CoA synthetase, alpha subunit |
| Dgeo_1090 | Dgeo_1090 | 1159883 | 1160698 | 816 | + | PROTEIN:transposase, IS4 |
| Dgeo_1091 | Dgeo_1091 | 1161105 | 1161362 | 258 | + | PROTEIN:hypothetical protein |
| Dgeo_1092 | Dgeo_1092 | 1161656 | 1163311 | 1656 | + | PROTEIN:hypothetical protein |
| Dgeo_1093 | Dgeo_1093 | 1163358 | 1163693 | 336 | + | PROTEIN:hypothetical protein |
| Dgeo_1094 | Dgeo_1094 | 1163876 | 1164199 | 324 | + | PROTEIN:hypothetical protein |
| Dgeo_1095 | Dgeo_1095 | 1164401 | 1165399 | 999 | + | PROTEIN:putative transposase |
| Dgeo_1097 | Dgeo_1097 | 1165746 | 1166099 | 354 | + | PROTEIN:phage integrase |
| Dgeo_1098 | Dgeo_1098 | 1166194 | 1166763 | 570 | + | PROTEIN:hypothetical protein |
| Dgeo_1099 | Dgeo_1099 | 1166773 | 1167915 | 1143 | - | PROTEIN:glycosyl transferase, group 1 |
| Dgeo_1100 | Dgeo_1100 | 1168003 | 1168848 | 846 | + | PROTEIN:degV family protein |
| Dgeo_1101 | Dgeo_1101 | 1169134 | 1172409 | 3276 | - | PROTEIN:isoleucyl-tRNA synthetase |
| Dgeo_1102 | Dgeo_1102 | 1172882 | 1173622 | 741 | + | PROTEIN:putative translaldolase |
| Dgeo_1103 | rho | 1173619 | 1174896 | 1278 | + | PROTEIN:transcription termination factor Rho |
| Dgeo_1104 | Dgeo_1104 | 1175007 | 1176341 | 1335 | + | PROTEIN:peptidase M23B |
| Dgeo_1105 | Dgeo_1105 | 1176338 | 1176703 | 366 | + | PROTEIN:response regulator receiver protein |
| Dgeo_1106 | Dgeo_1106 | 1176746 | 1177657 | 912 | + | PROTEIN:pyridoxine biosynthesis protein |
| Dgeo_1107 | Dgeo_1107 | 1177654 | 1178259 | 606 | + | PROTEIN:glutamine amidotransferase subunit PdxT |
| Dgeo_1108 | Dgeo_1108 | 1178285 | 1179025 | 741 | + | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_1109 | Dgeo_1109 | 1179022 | 1179768 | 747 | + | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_1110 | Dgeo_1110 | 1179765 | 1180967 | 1203 | - | PROTEIN:oxidoreductase-like protein |
| Dgeo_1111 | Dgeo_1111 | 1180964 | 1181851 | 888 | - | PROTEIN:NLP/P60 |
| Dgeo_1112 | Dgeo_1112 | 1181867 | 1182850 | 984 | - | PROTEIN:peptidase M19, renal dipeptidase |
| Dgeo_1113 | Dgeo_1113 | 1182856 | 1183542 | 687 | - | PROTEIN:HAD family hydrolase |
| Dgeo_1114 | Dgeo_1114 | 1183743 | 1184897 | 1155 | + | PROTEIN:Serine--pyruvate transaminase |
| Dgeo_1115 | Dgeo_1115 | 1184910 | 1185560 | 651 | - | PROTEIN:metallophosphoesterase |
| Dgeo_1116 | Dgeo_1116 | 1185589 | 1187085 | 1497 | + | PROTEIN:apolipoprotein N-acyltransferase |
| Dgeo_1117 | Dgeo_1117 | 1187014 | 1188123 | 1110 | - | PROTEIN:phage integrase |
| Dgeo_1118 | Dgeo_1118 | 1188202 | 1189164 | 963 | - | PROTEIN:diacylglycerol kinase, catalytic region |
| Dgeo_1119 | Dgeo_1119 | 1189192 | 1189560 | 369 | + | PROTEIN:cell surface receptor IPT/TIG |
| Dgeo_1120 | Dgeo_1120 | 1189583 | 1191040 | 1458 | - | PROTEIN:aldehyde dehydrogenase |
| Dgeo_1121 | hisS | 1191131 | 1192426 | 1296 | + | PROTEIN:histidyl-tRNA synthetase |
| Dgeo_1122 | aspS | 1192441 | 1194174 | 1734 | + | PROTEIN:aspartyl-tRNA synthetase |
| Dgeo_1123 | Dgeo_1123 | 1194595 | 1194870 | 276 | - | PROTEIN:phosphoglycerate mutase |
| Dgeo_1124 | Dgeo_1124 | 1194925 | 1196775 | 1851 | + | PROTEIN:excinuclease ABC, C subunit |
| Dgeo_1125 | Dgeo_1125 | 1197132 | 1197869 | 738 | + | PROTEIN:phosphoesterase, PA-phosphatase related |
| Dgeo_1127 | Dgeo_1127 | 1198797 | 1200209 | 1413 | - | PROTEIN:aspartate kinase |
| Dgeo_1128 | Dgeo_1128 | 1200270 | 1200617 | 348 | - | PROTEIN:putative copper resistance protein D |
| Dgeo_1129 | Dgeo_1129 | 1200605 | 1201057 | 453 | - | PROTEIN:putative copper resistance protein D |
| Dgeo_1130 | Dgeo_1130 | 1201062 | 1201460 | 399 | - | PROTEIN:copper resistance protein CopC |
| Dgeo_1131 | Dgeo_1131 | 1201510 | 1201959 | 450 | - | PROTEIN:nuclear export factor GLE1 |
| Dgeo_1132 | Dgeo_1132 | 1202034 | 1202387 | 354 | - | PROTEIN:hypothetical protein |
| Dgeo_1133 | Dgeo_1133 | 1202547 | 1203542 | 996 | + | PROTEIN:glyceraldehyde-3-phosphate dehydrogenase, |
| Dgeo_1134 | pgk | 1203628 | 1204797 | 1170 | + | PROTEIN:phosphoglycerate kinase |
| Dgeo_1135 | Dgeo_1135 | 1204809 | 1205543 | 735 | + | PROTEIN:triosephosphate isomerase |
| Dgeo_1136 | asnC | 1205676 | 1207025 | 1350 | + | PROTEIN:asparaginyl-tRNA synthetase |
| Dgeo_1137 | Dgeo_1137 | 1207143 | 1208030 | 888 | + | PROTEIN:phenazine biosynthesis PhzC/PhzF protein |
| Dgeo_1138 | Dgeo_1138 | 1208060 | 1208857 | 798 | + | PROTEIN:Cof protein |
| Dgeo_1139 | Dgeo_1139 | 1208919 | 1211249 | 2331 | + | PROTEIN:ATP-dependent DNA helicase RecG |
| Dgeo_1140 | Dgeo_1140 | 1211425 | 1212258 | 834 | + | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_1141 | Dgeo_1141 | 1212264 | 1213859 | 1596 | - | PROTEIN:gamma-glutamyltransferase |
| Dgeo_1142 | Dgeo_1142 | 1214013 | 1214945 | 933 | - | PROTEIN:PfkB |
| Dgeo_1143 | ksgA | 1215039 | 1215884 | 846 | - | PROTEIN:dimethyladenosine transferase |
| Dgeo_1144 | Dgeo_1144 | 1215961 | 1216809 | 849 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_1145 | Dgeo_1145 | 1216811 | 1218298 | 1488 | + | PROTEIN:YjeF-related protein-like protein |
| Dgeo_1146 | Dgeo_1146 | 1218308 | 1219012 | 705 | + | PROTEIN:hypothetical protein |
| Dgeo_1147 | Dgeo_1147 | 1219060 | 1221168 | 2109 | - | PROTEIN:EmrB/QacA family drug resistance transport |
| Dgeo_1148 | Dgeo_1148 | 1221165 | 1221659 | 495 | - | PROTEIN:MarR family transcriptional regulator |
| Dgeo_1149 | Dgeo_1149 | 1221926 | 1223962 | 2037 | + | PROTEIN:Pyrrolo-quinoline quinone |
| Dgeo_1150 | Dgeo_1150 | 1223966 | 1224601 | 636 | + | PROTEIN:hypothetical protein |
| Dgeo_1151 | Dgeo_1151 | 1224678 | 1225556 | 879 | - | PROTEIN:lysine biosynthesis enzyme LysX |
| Dgeo_1152 | Dgeo_1152 | 1225621 | 1225794 | 174 | - | PROTEIN:lysine biosynthesis protein LysW |
| Dgeo_1153 | Dgeo_1153 | 1225862 | 1226194 | 333 | - | PROTEIN:zinc finger, DHP-type |
| Dgeo_1154 | Dgeo_1154 | 1226204 | 1226713 | 510 | - | PROTEIN:3-isopropylmalate dehydratase small subuni |
| Dgeo_1155 | Dgeo_1155 | 1226738 | 1227445 | 708 | - | PROTEIN:hypothetical protein |
| Dgeo_1156 | Dgeo_1156 | 1227446 | 1228705 | 1260 | - | PROTEIN:3-isopropylmalate dehydratase large subuni |
| Dgeo_1157 | Dgeo_1157 | 1228732 | 1229319 | 588 | + | PROTEIN:TrkA-C |
| Dgeo_1158 | Dgeo_1158 | 1229499 | 1230665 | 1167 | + | PROTEIN:sodium/hydrogen exchanger |
| Dgeo_1159 | Dgeo_1159 | 1230738 | 1232270 | 1533 | - | PROTEIN:radical SAM family protein |
| Dgeo_1160 | Dgeo_1160 | 1232383 | 1232991 | 609 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_1161 | Dgeo_1161 | 1233125 | 1233352 | 228 | - | PROTEIN:hypothetical protein |
| Dgeo_1162 | Dgeo_1162 | 1233396 | 1234934 | 1539 | + | PROTEIN:L-aspartate oxidase |
| Dgeo_1163 | Dgeo_1163 | 1234975 | 1235811 | 837 | + | PROTEIN:nicotinate-nucleotide pyrophosphorylase |
| Dgeo_1164 | Dgeo_1164 | 1235814 | 1236809 | 996 | + | PROTEIN:quinolinate synthetase |
| Dgeo_1165 | Dgeo_1165 | 1236906 | 1238078 | 1173 | - | PROTEIN:hypothetical protein |
| Dgeo_1166 | Dgeo_1166 | 1238230 | 1239492 | 1263 | - | PROTEIN:isocitrate/isopropylmalate dehydrogenase |
| Dgeo_1167 | Dgeo_1167 | 1239688 | 1239957 | 270 | + | PROTEIN:hypothetical protein |
| Dgeo_1168 | Dgeo_1168 | 1240047 | 1240979 | 933 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1169 | Dgeo_1169 | 1240972 | 1241892 | 921 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1170 | Dgeo_1170 | 1241889 | 1243262 | 1374 | + | PROTEIN:hypothetical protein |
| Dgeo_1171 | Dgeo_1171 | 1243259 | 1244035 | 777 | + | PROTEIN:hypothetical protein |
| Dgeo_1172 | Dgeo_1172 | 1244040 | 1245533 | 1494 | + | PROTEIN:hypothetical protein |
| Dgeo_1173 | Dgeo_1173 | 1245673 | 1246569 | 897 | + | PROTEIN:ROK domain-containing protein |
| Dgeo_1174 | Dgeo_1174 | 1246690 | 1248267 | 1578 | + | PROTEIN:1-pyrroline-5-carboxylate dehydrogenase |
| Dgeo_1175 | Dgeo_1175 | 1248315 | 1248821 | 507 | + | PROTEIN:hypothetical protein |
| Dgeo_1176 | Dgeo_1176 | 1248815 | 1249699 | 885 | + | PROTEIN:hypothetical protein |
| Dgeo_1177 | Dgeo_1177 | 1249730 | 1251064 | 1335 | + | PROTEIN:aminotransferase class-III |
| Dgeo_1178 | Dgeo_1178 | 1251231 | 1251617 | 387 | - | PROTEIN:hypothetical protein |
| Dgeo_1179 | Dgeo_1179 | 1251841 | 1253019 | 1179 | - | PROTEIN:acyl-CoA dehydrogenase-like protein |
| Dgeo_1180 | Dgeo_1180 | 1253078 | 1253377 | 300 | - | PROTEIN:hypothetical protein |
| Dgeo_1181 | Dgeo_1181 | 1253390 | 1253692 | 303 | + | PROTEIN:hypothetical protein |
| Dgeo_1182 | Dgeo_1182 | 1253784 | 1254212 | 429 | + | PROTEIN:beta-hydroxyacyl-(acyl-carrier-protein) de |
| Dgeo_1183 | Dgeo_1183 | 1254328 | 1255473 | 1146 | + | PROTEIN:permease YjgP/YjgQ |
| Dgeo_1184 | Dgeo_1184 | 1255473 | 1256615 | 1143 | + | PROTEIN:monogalactosyldiacylglycerol synthase |
| Dgeo_1185 | Dgeo_1185 | 1256612 | 1257841 | 1230 | + | PROTEIN:polysaccharide deacetylase |
| Dgeo_1186 | Dgeo_1186 | 1257826 | 1258614 | 789 | + | PROTEIN:methyltransferase type 11 |
| Dgeo_1187 | Dgeo_1187 | 1258716 | 1259252 | 537 | - | PROTEIN:hypothetical protein |
| Dgeo_1188 | Dgeo_1188 | 1259293 | 1261380 | 2088 | + | PROTEIN:peptidase S8 and S53, subtilisin, kexin, s |
| Dgeo_1189 | Dgeo_1189 | 1261508 | 1263085 | 1578 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_1190 | Dgeo_1190 | 1263176 | 1264213 | 1038 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1191 | Dgeo_1191 | 1264290 | 1265291 | 1002 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1192 | Dgeo_1192 | 1265307 | 1266095 | 789 | - | PROTEIN:hypothetical protein |
| Dgeo_1193 | Dgeo_1193 | 1266188 | 1267315 | 1128 | - | PROTEIN:AlgP-related protein |
| Dgeo_1194 | Dgeo_1194 | 1267492 | 1269159 | 1668 | + | PROTEIN:DNA repair protein RecN |
| Dgeo_1195 | Dgeo_1195 | 1269244 | 1270326 | 1083 | + | PROTEIN:hypothetical protein |
| Dgeo_1196 | Dgeo_1196 | 1270534 | 1272123 | 1590 | + | PROTEIN:phosphoenolpyruvate carboxykinase |
| Dgeo_1197 | Dgeo_1197 | 1272241 | 1273128 | 888 | - | PROTEIN:hypothetical protein |
| Dgeo_1198 | Dgeo_1198 | 1273197 | 1275725 | 2529 | - | PROTEIN:Alpha-glucan phosphorylase |
| Dgeo_1199 | Dgeo_1199 | 1275837 | 1276133 | 297 | - | PROTEIN:hypothetical protein |
| Dgeo_1200 | Dgeo_1200 | 1276138 | 1276554 | 417 | - | PROTEIN:hypothetical protein |
| Dgeo_1201 | Dgeo_1201 | 1276719 | 1277918 | 1200 | - | PROTEIN:threonyl/alanyl tRNA synthetase, SAD |
| Dgeo_1202 | Dgeo_1202 | 1277915 | 1278715 | 801 | - | PROTEIN:aminoglycoside 3-N-acetyltransferase |
| Dgeo_1203 | Dgeo_1203 | 1278845 | 1281001 | 2157 | - | PROTEIN:glutamine synthetase, catalytic region |
| Dgeo_1204 | Dgeo_1204 | 1281265 | 1282623 | 1359 | + | PROTEIN:glutamine synthetase, type I |
| Dgeo_1205 | Dgeo_1205 | 1282878 | 1284032 | 1155 | + | PROTEIN:extracellular ligand-binding receptor |
| Dgeo_1206 | Dgeo_1206 | 1284277 | 1285290 | 1014 | + | PROTEIN:inner-membrane translocator |
| Dgeo_1207 | Dgeo_1207 | 1285287 | 1286912 | 1626 | + | PROTEIN:inner-membrane translocator |
| Dgeo_1208 | Dgeo_1208 | 1286909 | 1287721 | 813 | + | PROTEIN:ABC transporter related |
| Dgeo_1209 | Dgeo_1209 | 1287714 | 1288484 | 771 | + | PROTEIN:ABC transporter related |
| Dgeo_1210 | Dgeo_1210 | 1288683 | 1291268 | 2586 | + | PROTEIN:ATPase AAA-2 |
| Dgeo_1211 | Dgeo_1211 | 1291340 | 1291552 | 213 | - | PROTEIN:hypothetical protein |
| Dgeo_1212 | Dgeo_1212 | 1291648 | 1292997 | 1350 | - | PROTEIN:DNA repair protein RadA |
| Dgeo_1213 | Dgeo_1213 | 1292990 | 1293304 | 315 | - | PROTEIN:hypothetical protein |
| Dgeo_1214 | Dgeo_1214 | 1293321 | 1293656 | 336 | - | PROTEIN:hypothetical protein |
| Dgeo_1215 | Dgeo_1215 | 1293731 | 1295968 | 2238 | - | PROTEIN:ATPase AAA-2 |
| Dgeo_1216 | Dgeo_1216 | 1296105 | 1297421 | 1317 | - | PROTEIN:peptidase S41 |
| Dgeo_1217 | Dgeo_1217 | 1297494 | 1298267 | 774 | - | PROTEIN:5-carboxymethyl-2-hydroxymuconate delta-is |
| Dgeo_1218 | Dgeo_1218 | 1298304 | 1298888 | 585 | + | PROTEIN:NUDIX hydrolase |
| Dgeo_1219 | Dgeo_1219 | 1298913 | 1300391 | 1479 | + | PROTEIN:aminotransferase class-III |
| Dgeo_1220 | Dgeo_1220 | 1300401 | 1300907 | 507 | - | PROTEIN:hypothetical protein |
| Dgeo_1221 | Dgeo_1221 | 1300958 | 1302358 | 1401 | + | PROTEIN:acyl-CoA dehydrogenase-like protein |
| Dgeo_1222 | Dgeo_1222 | 1302509 | 1303444 | 936 | + | PROTEIN:hypothetical protein |
| Dgeo_1223 | Dgeo_1223 | 1303692 | 1305128 | 1437 | + | PROTEIN:major facilitator transporter |
| Dgeo_1224 | Dgeo_1224 | 1305136 | 1305741 | 606 | - | PROTEIN:NAD-dependent epimerase/dehydratase |
| Dgeo_1225 | Dgeo_1225 | 1305806 | 1306252 | 447 | - | PROTEIN:BadM/Rrf2 family transcriptional regulator |
| Dgeo_1226 | Dgeo_1226 | 1306355 | 1306942 | 588 | - | PROTEIN:HRDC |
| Dgeo_1228 | Dgeo_1228 | 1307576 | 1308262 | 687 | - | PROTEIN:hypothetical protein |
| Dgeo_1229 | Dgeo_1229 | 1308319 | 1309347 | 1029 | - | PROTEIN:zinc-binding alcohol dehydrogenase |
| Dgeo_1230 | Dgeo_1230 | 1309444 | 1309890 | 447 | - | PROTEIN:hypothetical protein |
| Dgeo_1231 | Dgeo_1231 | 1309989 | 1311143 | 1155 | + | PROTEIN:Acyl-CoA dehydrogenase, type 2-like protei |
| Dgeo_1232 | Dgeo_1232 | 1311205 | 1312338 | 1134 | + | PROTEIN:cell envelope-related transcriptional atte |
| Dgeo_1233 | Dgeo_1233 | 1312503 | 1312970 | 468 | + | PROTEIN:hypothetical protein |
| Dgeo_1234 | nusA | 1312999 | 1314186 | 1188 | + | PROTEIN:transcription elongation factor NusA |
| Dgeo_1235 | Dgeo_1235 | 1314196 | 1314477 | 282 | + | PROTEIN:hypothetical protein |
| Dgeo_1236 | infB | 1314546 | 1316351 | 1806 | + | PROTEIN:translation initiation factor IF-2 |
| Dgeo_1237 | Dgeo_1237 | 1316428 | 1317180 | 753 | - | PROTEIN:hypothetical protein |
| Dgeo_1238 | Dgeo_1238 | 1317493 | 1317855 | 363 | + | PROTEIN:hypothetical protein |
| Dgeo_1239 | Dgeo_1239 | 1318010 | 1320298 | 2289 | + | PROTEIN:bifunctional preprotein translocase subuni |
| Dgeo_1240 | Dgeo_1240 | 1320430 | 1322124 | 1695 | + | PROTEIN:dihydroxy-acid dehydratase |
| Dgeo_1241 | Dgeo_1241 | 1322291 | 1322974 | 684 | + | PROTEIN:cytochrome c biogenesis protein, transmemb |
| Dgeo_1242 | Dgeo_1242 | 1322959 | 1323672 | 714 | + | PROTEIN:ABC transporter related |
| Dgeo_1243 | Dgeo_1243 | 1323662 | 1324423 | 762 | + | PROTEIN:cytochrome c-type biogenesis protein CcmB |
| Dgeo_1244 | Dgeo_1244 | 1324452 | 1325165 | 714 | - | PROTEIN:hypothetical protein |
| Dgeo_1245 | Dgeo_1245 | 1325364 | 1326074 | 711 | + | PROTEIN:cytochrome c assembly protein |
| Dgeo_1246 | Dgeo_1246 | 1326183 | 1326650 | 468 | + | PROTEIN:cytochrome c-type biogenesis protein CcmE |
| Dgeo_1247 | Dgeo_1247 | 1326661 | 1328643 | 1983 | + | PROTEIN:cytochrome c assembly protein |
| Dgeo_1248 | Dgeo_1248 | 1328664 | 1329242 | 579 | + | PROTEIN:redoxin |
| Dgeo_1249 | Dgeo_1249 | 1329239 | 1329757 | 519 | + | PROTEIN:cytochrome C biogenesis protein |
| Dgeo_1250 | Dgeo_1250 | 1329754 | 1330791 | 1038 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_1251 | Dgeo_1251 | 1330788 | 1331351 | 564 | + | PROTEIN:Rieske (2Fe-2S) region |
| Dgeo_1252 | Dgeo_1252 | 1331379 | 1332482 | 1104 | - | PROTEIN:luciferase-like protein |
| Dgeo_1253 | Dgeo_1253 | 1332671 | 1333534 | 864 | + | PROTEIN:Citryl-CoA lyase |
| Dgeo_1254 | Dgeo_1254 | 1333531 | 1334016 | 486 | + | PROTEIN:MaoC-like protein dehydratase |
| Dgeo_1255 | Dgeo_1255 | 1334044 | 1334697 | 654 | - | PROTEIN:putative oxidoreductase |
| Dgeo_1256 | Dgeo_1256 | 1334719 | 1335021 | 303 | - | PROTEIN:D-threo-aldose 1-dehydrogenase |
| Dgeo_1257 | Dgeo_1257 | 1335183 | 1336352 | 1170 | + | PROTEIN:homocitrate synthase |
| Dgeo_1258 | Dgeo_1258 | 1336414 | 1337229 | 816 | + | PROTEIN:hypothetical protein |
| Dgeo_1259 | Dgeo_1259 | 1337325 | 1338407 | 1083 | + | PROTEIN:benzoate transporter |
| Dgeo_1260 | Dgeo_1260 | 1338440 | 1339072 | 633 | + | PROTEIN:hypothetical protein |
| Dgeo_1261 | Dgeo_1261 | 1339111 | 1340304 | 1194 | + | PROTEIN:cysteine desulphurase-like protein |
| Dgeo_1262 | Dgeo_1262 | 1340382 | 1341041 | 660 | - | PROTEIN:hypothetical protein |
| Dgeo_1263 | Dgeo_1263 | 1341154 | 1343190 | 2037 | + | PROTEIN:hypothetical protein |
| Dgeo_1264 | Dgeo_1264 | 1343228 | 1344196 | 969 | - | PROTEIN:LmbE-like protein protein |
| Dgeo_1265 | Dgeo_1265 | 1344196 | 1344882 | 687 | - | PROTEIN:ribose 5-phosphate isomerase |
| Dgeo_1266 | Dgeo_1266 | 1344944 | 1346173 | 1230 | + | PROTEIN:deoxyribose-phosphate aldolase |
| Dgeo_1267 | Dgeo_1267 | 1346184 | 1346747 | 564 | + | PROTEIN:maf protein |
| Dgeo_1268 | Dgeo_1268 | 1346793 | 1347191 | 399 | - | PROTEIN:OsmC-like protein protein |
| Dgeo_1269 | Dgeo_1269 | 1347211 | 1348461 | 1251 | - | PROTEIN:response regulator receiver sensor signal |
| Dgeo_1270 | Dgeo_1270 | 1348458 | 1351283 | 2826 | - | PROTEIN:PAS/PAC sensor signal transduction histidi |
| Dgeo_1271 | Dgeo_1271 | 1351688 | 1352770 | 1083 | - | PROTEIN:shikimate 5-dehydrogenase |
| Dgeo_1272 | Dgeo_1272 | 1352772 | 1353830 | 1059 | + | PROTEIN:PhoH-like protein protein |
| Dgeo_1273 | Dgeo_1273 | 1353916 | 1354383 | 468 | + | PROTEIN:hypothetical protein |
| Dgeo_1274 | Dgeo_1274 | 1354380 | 1354757 | 378 | + | PROTEIN:diacylglycerol kinase |
| Dgeo_1275 | Dgeo_1275 | 1354792 | 1355484 | 693 | + | PROTEIN:Crp/FNR family transcriptional regulator |
| Dgeo_1276 | Dgeo_1276 | 1355566 | 1357122 | 1557 | + | PROTEIN:hypothetical protein |
| Dgeo_1277 | Dgeo_1277 | 1357142 | 1357702 | 561 | - | PROTEIN:hypothetical protein |
| Dgeo_1278 | Dgeo_1278 | 1357763 | 1360306 | 2544 | - | PROTEIN:peptidase U32 |
| Dgeo_1279 | Dgeo_1279 | 1360359 | 1360835 | 477 | - | PROTEIN:hypothetical protein |
| Dgeo_1280 | Dgeo_1280 | 1361286 | 1361798 | 513 | + | PROTEIN:rhodanese-like protein |
| Dgeo_1281 | Dgeo_1281 | 1361798 | 1362154 | 357 | + | PROTEIN:hypothetical protein |
| Dgeo_1282 | Dgeo_1282 | 1362274 | 1362954 | 681 | - | PROTEIN:peptidase, membrane zinc metallopeptidase, |
| Dgeo_1283 | Dgeo_1283 | 1363076 | 1363366 | 291 | - | PROTEIN:stage V sporulation protein S |
| Dgeo_1284 | Dgeo_1284 | 1363505 | 1364284 | 780 | - | PROTEIN:methionine aminopeptidase, type I |
| Dgeo_1285 | Dgeo_1285 | 1364638 | 1365978 | 1341 | + | PROTEIN:hypothetical protein |
| Dgeo_1286 | Dgeo_1286 | 1365971 | 1366420 | 450 | + | PROTEIN:cytidine deaminase, homotetrameric |
| Dgeo_1287 | Dgeo_1287 | 1366534 | 1367865 | 1332 | - | PROTEIN:isocitrate lyase |
| Dgeo_1288 | Dgeo_1288 | 1368112 | 1368750 | 639 | - | PROTEIN:HAD family hydrolase |
| Dgeo_1289 | Dgeo_1289 | 1368822 | 1369520 | 699 | - | PROTEIN:hypothetical protein |
| Dgeo_1290 | Dgeo_1290 | 1369600 | 1370607 | 1008 | + | PROTEIN:phosphoesterase, RecJ-like protein |
| Dgeo_1291 | Dgeo_1291 | 1370612 | 1371775 | 1164 | - | PROTEIN:beta-lactamase, putative |
| Dgeo_1292 | Dgeo_1292 | 1371779 | 1372639 | 861 | - | PROTEIN:degV family protein |
| Dgeo_1293 | Dgeo_1293 | 1372665 | 1373609 | 945 | - | PROTEIN:diacylglycerol kinase, catalytic region |
| Dgeo_1294 | Dgeo_1294 | 1373633 | 1374790 | 1158 | - | PROTEIN:UDP-N-acetylglucosamine 2-epimerase |
| Dgeo_1295 | Dgeo_1295 | 1374905 | 1376116 | 1212 | - | PROTEIN:glycosyl transferase family protein |
| Dgeo_1296 | Dgeo_1296 | 1376123 | 1376746 | 624 | - | PROTEIN:uracil phosphoribosyltransferase |
| Dgeo_1297 | Dgeo_1297 | 1376861 | 1377712 | 852 | - | PROTEIN:aminoglycoside phosphotransferase |
| Dgeo_1298 | Dgeo_1298 | 1377730 | 1378755 | 1026 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_1299 | Dgeo_1299 | 1378823 | 1379533 | 711 | + | PROTEIN:hypothetical protein |
| Dgeo_1300 | Dgeo_1300 | 1379598 | 1380347 | 750 | + | PROTEIN:hypothetical protein |
| Dgeo_1301 | Dgeo_1301 | 1380344 | 1381228 | 885 | + | PROTEIN:hypothetical protein |
| Dgeo_1302 | Dgeo_1302 | 1381407 | 1382597 | 1191 | - | PROTEIN:IS4 family transposase |
| Dgeo_1303 | Dgeo_1303 | 1382794 | 1384056 | 1263 | - | PROTEIN:hypothetical protein |
| Dgeo_1304 | Dgeo_1304 | 1384152 | 1386101 | 1950 | - | PROTEIN:glucose-6-phosphate isomerase |
| Dgeo_1305 | Dgeo_1305 | 1386136 | 1387380 | 1245 | + | PROTEIN:Acetyl-CoA C-acetyltransferase |
| Dgeo_1306 | Dgeo_1306 | 1387377 | 1388738 | 1362 | + | PROTEIN:3-oxoacid CoA-transferase, subunit B |
| Dgeo_1307 | Dgeo_1307 | 1388824 | 1389528 | 705 | - | PROTEIN:TetR family transcriptional regulator |
| Dgeo_1308 | Dgeo_1308 | 1389577 | 1391877 | 2301 | + | PROTEIN:(p)ppGpp synthetase I, SpoT/RelA |
| Dgeo_1309 | Dgeo_1309 | 1391893 | 1392888 | 996 | - | PROTEIN:Ion transport 2 |
| Dgeo_1310 | Dgeo_1310 | 1392943 | 1393662 | 720 | - | PROTEIN:hypothetical protein |
| Dgeo_1311 | Dgeo_1311 | 1393694 | 1393897 | 204 | - | PROTEIN:regulatory protein, MerR |
| Dgeo_1312 | Dgeo_1312 | 1394250 | 1395020 | 771 | + | PROTEIN:hypothetical protein |
| Dgeo_1313 | Dgeo_1313 | 1395084 | 1396298 | 1215 | - | PROTEIN:hypothetical protein |
| Dgeo_1314 | Dgeo_1314 | 1396279 | 1396587 | 309 | - | PROTEIN:ArsR family transcriptional regulator |
| Dgeo_1315 | Dgeo_1315 | 1396606 | 1397568 | 963 | - | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_1316 | Dgeo_1316 | 1397861 | 1399141 | 1281 | + | PROTEIN:S-layer-like protein array-related protein |
| Dgeo_1317 | Dgeo_1317 | 1399351 | 1400577 | 1227 | - | PROTEIN:carboxynorspermidine decarboxylase |
| Dgeo_1318 | Dgeo_1318 | 1400641 | 1401366 | 726 | - | PROTEIN:2-phosphosulfolactate phosphatase |
| Dgeo_1319 | Dgeo_1319 | 1401363 | 1402052 | 690 | - | PROTEIN:ribulose-phosphate 3-epimerase |
| Dgeo_1320 | Dgeo_1320 | 1402217 | 1404385 | 2169 | + | PROTEIN:methylmalonyl-CoA mutase |
| Dgeo_1321 | Dgeo_1321 | 1404474 | 1404962 | 489 | + | PROTEIN:hypothetical protein |
| Dgeo_1322 | Dgeo_1322 | 1404988 | 1406043 | 1056 | + | PROTEIN:arginine/ornithine transport system ATPase |
| Dgeo_1323 | Dgeo_1323 | 1406250 | 1407173 | 924 | - | PROTEIN:dessication-associated protein |
| Dgeo_1324 | Dgeo_1324 | 1407608 | 1408543 | 936 | - | PROTEIN:bifunctional nicotinamide mononucleotide a |
| Dgeo_1325 | Dgeo_1325 | 1408540 | 1410000 | 1461 | - | PROTEIN:nicotinate phosphoribosyltransferase |
| Dgeo_1326 | Dgeo_1326 | 1409997 | 1411391 | 1395 | - | PROTEIN:major facilitator transporter |
| Dgeo_1327 | nadE | 1411465 | 1412331 | 867 | + | PROTEIN:NAD synthetase |
| Dgeo_1328 | Dgeo_1328 | 1412420 | 1413391 | 972 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_1329 | Dgeo_1329 | 1413461 | 1413805 | 345 | + | PROTEIN:histidine triad (HIT) protein |
| Dgeo_1330 | Dgeo_1330 | 1413802 | 1414494 | 693 | + | PROTEIN:HAD family hydrolase |
| Dgeo_1331 | Dgeo_1331 | 1414504 | 1415394 | 891 | - | PROTEIN:L-serine dehydratase, iron-sulfur-dependen |
| Dgeo_1332 | Dgeo_1332 | 1415416 | 1416081 | 666 | - | PROTEIN:L-serine dehydratase, iron-sulfur-dependen |
| Dgeo_R0026 | Dgeo_R0026 | 1416271 | 1416345 | 75 | - | RNA:Val tRNA |
| Dgeo_1333 | Dgeo_1333 | 1416399 | 1417805 | 1407 | - | PROTEIN:amidase |
| Dgeo_1334 | Dgeo_1334 | 1417889 | 1418800 | 912 | + | PROTEIN:prephenate dehydratase |
| Dgeo_1335 | Dgeo_1335 | 1418840 | 1419118 | 279 | - | PROTEIN:hypothetical protein |
| Dgeo_1336 | metH | 1419459 | 1423139 | 3681 | + | PROTEIN:B12-dependent methionine synthase |
| Dgeo_1337 | Dgeo_1337 | 1423136 | 1423876 | 741 | + | PROTEIN:5,10-methylenetetrahydrofolate reductase-r |
| Dgeo_1339 | Dgeo_1339 | 1424743 | 1426041 | 1299 | - | PROTEIN:hypothetical protein |
| Dgeo_1340 | Dgeo_1340 | 1426160 | 1427188 | 1029 | - | PROTEIN:oligopeptide/dipeptide ABC transporter, AT |
| Dgeo_1341 | Dgeo_1341 | 1427185 | 1428234 | 1050 | - | PROTEIN:oligopeptide/dipeptide ABC transporter, AT |
| Dgeo_1342 | Dgeo_1342 | 1428324 | 1429466 | 1143 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1343 | Dgeo_1343 | 1429490 | 1430470 | 981 | - | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1344 | Dgeo_1344 | 1430551 | 1432467 | 1917 | - | PROTEIN:extracellular solute-binding protein |
| Dgeo_1345 | secG | 1432657 | 1432881 | 225 | - | PROTEIN:preprotein translocase subunit SecG |
| Dgeo_1346 | Dgeo_1346 | 1433106 | 1434317 | 1212 | - | PROTEIN:RNA polymerase, sigma 28 subunit |
| Dgeo_R0027 | Dgeo_R0027 | 1435008 | 1435090 | 83 | - | RNA:Leu tRNA |
| Dgeo_1347 | Dgeo_1347 | 1435158 | 1436411 | 1254 | - | PROTEIN:phosphoribosylamine--glycine ligase |
| Dgeo_1348 | Dgeo_1348 | 1436408 | 1437331 | 924 | - | PROTEIN:beta-lactamase-like protein |
| Dgeo_1349 | Dgeo_1349 | 1437328 | 1437501 | 174 | - | PROTEIN:hypothetical protein |
| Dgeo_1350 | Dgeo_1350 | 1437590 | 1437928 | 339 | - | PROTEIN:hypothetical protein |
| Dgeo_1351 | Dgeo_1351 | 1437928 | 1438410 | 483 | - | PROTEIN:hypothetical protein |
| Dgeo_1352 | Dgeo_1352 | 1438441 | 1439628 | 1188 | + | PROTEIN:amidase |
| Dgeo_1353 | Dgeo_1353 | 1439639 | 1440445 | 807 | + | PROTEIN:hypothetical protein |
| Dgeo_1354 | Dgeo_1354 | 1440500 | 1440985 | 486 | + | PROTEIN:hypothetical protein |
| Dgeo_1355 | Dgeo_1355 | 1440990 | 1441334 | 345 | - | PROTEIN:hypothetical protein |
| Dgeo_1356 | Dgeo_1356 | 1441383 | 1441598 | 216 | - | PROTEIN:hypothetical protein |
| Dgeo_1357 | Dgeo_1357 | 1441595 | 1441870 | 276 | - | PROTEIN:hypothetical protein |
| Dgeo_1358 | Dgeo_1358 | 1441875 | 1442177 | 303 | - | PROTEIN:hypothetical protein |
| Dgeo_1359 | Dgeo_1359 | 1442260 | 1443123 | 864 | - | PROTEIN:hypothetical protein |
| Dgeo_1360 | Dgeo_1360 | 1443191 | 1444447 | 1257 | - | PROTEIN:hypothetical protein |
| Dgeo_1361 | Dgeo_1361 | 1444444 | 1444728 | 285 | - | PROTEIN:hypothetical protein |
| Dgeo_1362 | Dgeo_1362 | 1445011 | 1445235 | 225 | - | PROTEIN:hypothetical protein |
| Dgeo_1363 | Dgeo_1363 | 1445447 | 1445641 | 195 | - | PROTEIN:hypothetical protein |
| Dgeo_1364 | Dgeo_1364 | 1445638 | 1445823 | 186 | - | PROTEIN:hypothetical protein |
| Dgeo_1365 | Dgeo_1365 | 1445921 | 1446163 | 243 | - | PROTEIN:hypothetical protein |
| Dgeo_1366 | Dgeo_1366 | 1446529 | 1447239 | 711 | + | PROTEIN:putative prophage repressor |
| Dgeo_1367 | Dgeo_1367 | 1447276 | 1447707 | 432 | + | PROTEIN:hypothetical protein |
| Dgeo_1368 | Dgeo_1368 | 1447734 | 1449095 | 1362 | + | PROTEIN:recombinase |
| Dgeo_1369 | Dgeo_1369 | 1449128 | 1450507 | 1380 | + | PROTEIN:tRNA-i(6)A37 modification enzyme MiaB |
| Dgeo_1370 | Dgeo_1370 | 1450619 | 1451584 | 966 | - | PROTEIN:periplasmic binding protein |
| Dgeo_1371 | Dgeo_1371 | 1451862 | 1452413 | 552 | + | PROTEIN:hypothetical protein |
| Dgeo_1372 | Dgeo_1372 | 1452702 | 1454048 | 1347 | - | PROTEIN:transposase IS66 |
| Dgeo_1373 | Dgeo_1373 | 1454150 | 1454530 | 381 | + | PROTEIN:hypothetical protein |
| Dgeo_1374 | Dgeo_1374 | 1454604 | 1456205 | 1602 | - | PROTEIN:ABC transporter related |
| Dgeo_1375 | Dgeo_1375 | 1456269 | 1457090 | 822 | + | PROTEIN:phosphoesterase, PA-phosphatase related |
| Dgeo_1376 | Dgeo_1376 | 1457170 | 1458330 | 1161 | + | PROTEIN:hypothetical protein |
| Dgeo_1377 | Dgeo_1377 | 1458330 | 1459028 | 699 | + | PROTEIN:ABC transporter related |
| Dgeo_1378 | Dgeo_1378 | 1459067 | 1459516 | 450 | - | PROTEIN:hypothetical protein |
| Dgeo_1379 | Dgeo_1379 | 1459607 | 1461556 | 1950 | - | PROTEIN:hypothetical protein |
| Dgeo_1380 | Dgeo_1380 | 1461598 | 1462665 | 1068 | + | PROTEIN:alanine racemase |
| Dgeo_1381 | Dgeo_1381 | 1462678 | 1463718 | 1041 | - | PROTEIN:isopentenyl pyrophosphate isomerase |
| Dgeo_R0028 | Dgeo_R0028 | 1463818 | 1463894 | 77 | + | RNA:Met tRNA |
| Dgeo_1382 | Dgeo_1382 | 1463972 | 1464346 | 375 | - | PROTEIN:hypothetical protein |
| Dgeo_1383 | Dgeo_1383 | 1464348 | 1465418 | 1071 | - | PROTEIN:butyrate kinase |
| Dgeo_1384 | Dgeo_1384 | 1465514 | 1466074 | 561 | - | PROTEIN:sigma 54 modulation protein/ribosomal prot |
| Dgeo_1385 | Dgeo_1385 | 1466747 | 1468549 | 1803 | + | PROTEIN:ABC transporter related |
| Dgeo_1386 | Dgeo_1386 | 1468680 | 1469963 | 1284 | - | PROTEIN:UDP-N-acetylglucosamine 1-carboxyvinyltran |
| Dgeo_1387 | Dgeo_1387 | 1470304 | 1470531 | 228 | + | PROTEIN:hypothetical protein |
| Dgeo_1388 | Dgeo_1388 | 1470617 | 1471528 | 912 | - | PROTEIN:NLP/P60 |
| Dgeo_1389 | Dgeo_1389 | 1471758 | 1473053 | 1296 | + | PROTEIN:major facilitator transporter |
| Dgeo_1390 | Dgeo_1390 | 1473302 | 1475047 | 1746 | - | PROTEIN:citrate transporter |
| Dgeo_1391 | Dgeo_1391 | 1475204 | 1476280 | 1077 | - | PROTEIN:acetyl-lysine deacetylase |
| Dgeo_1392 | Dgeo_1392 | 1476264 | 1477172 | 909 | - | PROTEIN:hypothetical protein |
| Dgeo_1393 | Dgeo_1393 | 1477309 | 1477599 | 291 | + | PROTEIN:hypothetical protein |
| Dgeo_1394 | Dgeo_1394 | 1477606 | 1479105 | 1500 | - | PROTEIN:Mg chelatase-related protein |
| Dgeo_1395 | Dgeo_1395 | 1479105 | 1479947 | 843 | - | PROTEIN:degV family protein |
| Dgeo_1396 | Dgeo_1396 | 1480075 | 1480389 | 315 | - | PROTEIN:twin arginine-targeting protein translocas |
| Dgeo_1397 | Dgeo_1397 | 1480486 | 1482417 | 1932 | - | PROTEIN:serine/threonine protein kinase |
| Dgeo_1398 | Dgeo_1398 | 1482509 | 1483918 | 1410 | + | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_1399 | Dgeo_1399 | 1483973 | 1484260 | 288 | - | PROTEIN:hypothetical protein |
| Dgeo_1400 | Dgeo_1400 | 1484279 | 1485484 | 1206 | - | PROTEIN:hypothetical protein |
| Dgeo_1401 | Dgeo_1401 | 1485497 | 1486852 | 1356 | + | PROTEIN:recombination factor protein RarA |
| Dgeo_1402 | Dgeo_1402 | 1486912 | 1487868 | 957 | - | PROTEIN:aldo/keto reductase |
| Dgeo_1404 | Dgeo_1404 | 1489913 | 1490734 | 822 | - | PROTEIN:orotidine 5'-phosphate decarboxylase subfa |
| Dgeo_1405 | Dgeo_1405 | 1490756 | 1491613 | 858 | + | PROTEIN:hypothetical protein |
| Dgeo_1406 | Dgeo_1406 | 1491684 | 1492151 | 468 | + | PROTEIN:BadM/Rrf2 family transcriptional regulator |
| Dgeo_1407 | Dgeo_1407 | 1492372 | 1493937 | 1566 | + | PROTEIN:ferredoxin--nitrite reductase |
| Dgeo_1408 | Dgeo_1408 | 1493934 | 1494479 | 546 | + | PROTEIN:adenylylsulfate kinase |
| Dgeo_1409 | Dgeo_1409 | 1494476 | 1495195 | 720 | + | PROTEIN:phosphoadenosine phosphosulfate reductase |
| Dgeo_1410 | sat | 1495278 | 1496447 | 1170 | + | PROTEIN:sulfate adenylyltransferase |
| Dgeo_1411 | Dgeo_1411 | 1496522 | 1497484 | 963 | + | PROTEIN:ABC transporter, substrate-binding protein |
| Dgeo_1412 | Dgeo_1412 | 1497489 | 1498322 | 834 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1413 | Dgeo_1413 | 1498319 | 1499062 | 744 | + | PROTEIN:ABC transporter related |
| Dgeo_1414 | Dgeo_1414 | 1499147 | 1500493 | 1347 | - | PROTEIN:transposase IS66 |
| Dgeo_1415 | Dgeo_1415 | 1500631 | 1502031 | 1401 | - | PROTEIN:MATE efflux family protein |
| Dgeo_1416 | Dgeo_1416 | 1502209 | 1503456 | 1248 | - | PROTEIN:acetylornithine and succinylornithine amin |
| Dgeo_1417 | Dgeo_1417 | 1503561 | 1505381 | 1821 | - | PROTEIN:hypothetical protein |
| Dgeo_1418 | Dgeo_1418 | 1505378 | 1506418 | 1041 | - | PROTEIN:hypothetical protein |
| Dgeo_1419 | Dgeo_1419 | 1506483 | 1507109 | 627 | - | PROTEIN:Crp/FNR family transcriptional regulator |
| Dgeo_1420 | Dgeo_1420 | 1507323 | 1508243 | 921 | - | PROTEIN:histone deacetylase superfamily protein |
| Dgeo_1421 | Dgeo_1421 | 1508432 | 1509286 | 855 | + | PROTEIN:PfkB |
| Dgeo_1422 | Dgeo_1422 | 1509303 | 1509653 | 351 | - | PROTEIN:aspartate alpha-decarboxylase |
| Dgeo_1423 | Dgeo_1423 | 1509748 | 1511301 | 1554 | - | PROTEIN:fibronectin-binding A-like protein |
| Dgeo_1424 | Dgeo_1424 | 1511414 | 1512253 | 840 | - | PROTEIN:MerR family transcriptional regulator |
| Dgeo_1425 | Dgeo_1425 | 1512360 | 1514891 | 2532 | - | PROTEIN:surface antigen (D15) |
| Dgeo_1426 | Dgeo_1426 | 1515115 | 1515774 | 660 | + | PROTEIN:hypothetical protein |
| Dgeo_1427 | Dgeo_1427 | 1515771 | 1516538 | 768 | + | PROTEIN:hypothetical protein |
| Dgeo_1428 | Dgeo_1428 | 1516581 | 1517669 | 1089 | - | PROTEIN:RelA/SpoT |
| Dgeo_1429 | Dgeo_1429 | 1517763 | 1518074 | 312 | + | PROTEIN:hypothetical protein |
| Dgeo_1430 | Dgeo_1430 | 1518088 | 1519512 | 1425 | + | PROTEIN:GntR family transcriptional regulator |
| Dgeo_1431 | Dgeo_1431 | 1519532 | 1519897 | 366 | + | PROTEIN:hypothetical protein |
| Dgeo_1432 | rpsT | 1519974 | 1520261 | 288 | - | PROTEIN:30S ribosomal protein S20 |
| Dgeo_1433 | Dgeo_1433 | 1520383 | 1520958 | 576 | + | PROTEIN:regulatory protein RecX |
| Dgeo_R0029 | Dgeo_R0029 | 1521063 | 1521139 | 77 | + | RNA:Pro tRNA |
| Dgeo_1434 | Dgeo_1434 | 1521206 | 1521937 | 732 | + | PROTEIN:metallophosphoesterase |
| Dgeo_1435 | Dgeo_1435 | 1522063 | 1522581 | 519 | + | PROTEIN:hypothetical protein |
| Dgeo_1436 | Dgeo_1436 | 1522658 | 1523731 | 1074 | + | PROTEIN:luciferase-like protein |
| Dgeo_1437 | Dgeo_1437 | 1523791 | 1524693 | 903 | + | PROTEIN:hypothetical protein |
| Dgeo_1438 | Dgeo_1438 | 1525086 | 1526414 | 1329 | + | PROTEIN:secretion protein HlyD |
| Dgeo_1439 | Dgeo_1439 | 1526411 | 1529782 | 3372 | + | PROTEIN:acriflavin resistance protein |
| Dgeo_R0030 | Dgeo_R0030 | 1530030 | 1530105 | 76 | - | RNA:Lys tRNA |
| Dgeo_1440 | Dgeo_1440 | 1530194 | 1531546 | 1353 | - | PROTEIN:signal recognition particle protein |
| Dgeo_1441 | Dgeo_1441 | 1531713 | 1532894 | 1182 | + | PROTEIN:Acetyl-CoA C-acetyltransferase |
| Dgeo_1442 | Dgeo_1442 | 1532935 | 1533252 | 318 | + | PROTEIN:YCII-related |
| Dgeo_1443 | Dgeo_1443 | 1533249 | 1533491 | 243 | + | PROTEIN:hypothetical protein |
| Dgeo_1444 | Dgeo_1444 | 1533488 | 1534324 | 837 | + | PROTEIN:3-hydroxybutyryl-CoA dehydrogenase |
| Dgeo_1445 | Dgeo_1445 | 1534399 | 1534659 | 261 | + | PROTEIN:hypothetical protein |
| Dgeo_1446 | Dgeo_1446 | 1534641 | 1535228 | 588 | - | PROTEIN:hypothetical protein |
| Dgeo_1447 | Dgeo_1447 | 1535317 | 1535814 | 498 | + | PROTEIN:hypothetical protein |
| Dgeo_1448 | Dgeo_1448 | 1535868 | 1536548 | 681 | - | PROTEIN:hypothetical protein |
| Dgeo_1449 | Dgeo_1449 | 1536560 | 1536964 | 405 | - | PROTEIN:hypothetical protein |
| Dgeo_1450 | Dgeo_1450 | 1536972 | 1537982 | 1011 | - | PROTEIN:abortive infection protein |
| Dgeo_1451 | Dgeo_1451 | 1538020 | 1539057 | 1038 | - | PROTEIN:threonine aldolase |
| Dgeo_1452 | Dgeo_1452 | 1539128 | 1540507 | 1380 | - | PROTEIN:hypothetical protein |
| Dgeo_1453 | fusA | 1540618 | 1542651 | 2034 | - | PROTEIN:elongation factor G |
| Dgeo_1454 | Dgeo_1454 | 1542955 | 1543821 | 867 | + | PROTEIN:hypothetical protein |
| Dgeo_1455 | Dgeo_1455 | 1543942 | 1544412 | 471 | + | PROTEIN:hypothetical protein |
| Dgeo_1456 | Dgeo_1456 | 1544452 | 1545129 | 678 | + | PROTEIN:hypothetical protein |
| Dgeo_1457 | Dgeo_1457 | 1545126 | 1546100 | 975 | + | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_1458 | Dgeo_1458 | 1546136 | 1547137 | 1002 | + | PROTEIN:3-isopropylmalate dehydrogenase |
| Dgeo_1459 | Dgeo_1459 | 1547196 | 1547651 | 456 | - | PROTEIN:hypothetical protein |
| Dgeo_1460 | Dgeo_1460 | 1547664 | 1548188 | 525 | - | PROTEIN:hypothetical protein |
| Dgeo_1461 | Dgeo_1461 | 1548468 | 1549382 | 915 | + | PROTEIN:phospho-N-acetylmuramoyl-pentapeptide-tran |
| Dgeo_1462 | Dgeo_1462 | 1549456 | 1550412 | 957 | + | PROTEIN:hypothetical protein |
| Dgeo_1463 | Dgeo_1463 | 1550423 | 1550644 | 222 | - | PROTEIN:hypothetical protein |
| Dgeo_1464 | Dgeo_1464 | 1550737 | 1551396 | 660 | + | PROTEIN:hypothetical protein |
| Dgeo_1465 | Dgeo_1465 | 1551592 | 1552431 | 840 | + | PROTEIN:hypothetical protein |
| Dgeo_1466 | Dgeo_1466 | 1552428 | 1552850 | 423 | + | PROTEIN:hypothetical protein |
| Dgeo_1467 | Dgeo_1467 | 1552954 | 1554567 | 1614 | + | PROTEIN:sensor protein |
| Dgeo_1468 | Dgeo_1468 | 1554716 | 1555771 | 1056 | - | PROTEIN:hypothetical protein |
| Dgeo_1469 | Dgeo_1469 | 1556072 | 1556752 | 681 | + | PROTEIN:cytochrome c, class I |
| Dgeo_1470 | Dgeo_1470 | 1556763 | 1557089 | 327 | - | PROTEIN:hypothetical protein |
| Dgeo_1471 | Dgeo_1471 | 1557223 | 1558191 | 969 | - | PROTEIN:hypothetical protein |
| Dgeo_1472 | Dgeo_1472 | 1558335 | 1558712 | 378 | + | PROTEIN:hypothetical protein |
| Dgeo_1473 | Dgeo_1473 | 1558770 | 1559480 | 711 | + | PROTEIN:late embryogenesis abundant protein |
| Dgeo_1474 | Dgeo_1474 | 1559586 | 1559927 | 342 | + | PROTEIN:ATP-dependent Clp protease adaptor protein |
| Dgeo_1475 | Dgeo_1475 | 1559938 | 1562151 | 2214 | + | PROTEIN:ATPase AAA-2 |
| Dgeo_1476 | Dgeo_1476 | 1562325 | 1563437 | 1113 | - | PROTEIN:oxidoreductase-like protein |
| Dgeo_1477 | Dgeo_1477 | 1563555 | 1564115 | 561 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_1478 | Dgeo_1478 | 1564112 | 1564732 | 621 | + | PROTEIN:hypothetical protein |
| Dgeo_1479 | Dgeo_1479 | 1564771 | 1566120 | 1350 | - | PROTEIN:carboxyl-terminal protease |
| Dgeo_1480 | Dgeo_1480 | 1566203 | 1566958 | 756 | + | PROTEIN:cell division ATP-binding protein FtsE |
| Dgeo_1481 | Dgeo_1481 | 1566948 | 1567886 | 939 | + | PROTEIN:hypothetical protein |
| Dgeo_1482 | Dgeo_1482 | 1567883 | 1569451 | 1569 | + | PROTEIN:peptidase M23B |
| Dgeo_1483 | rpsP | 1569646 | 1569894 | 249 | + | PROTEIN:30S ribosomal protein S16 |
| Dgeo_1484 | Dgeo_1484 | 1570027 | 1570275 | 249 | + | PROTEIN:hypothetical protein |
| Dgeo_1485 | Dgeo_1485 | 1570286 | 1570822 | 537 | + | PROTEIN:16S rRNA processing protein RimM |
| Dgeo_1486 | Dgeo_1486 | 1570851 | 1571648 | 798 | + | PROTEIN:tRNA (guanine-N1)-methyltransferase |
| Dgeo_1487 | Dgeo_1487 | 1571727 | 1572338 | 612 | + | PROTEIN:peptidase S16, lon-like protein |
| Dgeo_1488 | Dgeo_1488 | 1572380 | 1573270 | 891 | - | PROTEIN:formyltetrahydrofolate deformylase |
| Dgeo_1489 | Dgeo_1489 | 1573316 | 1574428 | 1113 | + | PROTEIN:peptidase M29, aminopeptidase II |
| Dgeo_1490 | Dgeo_1490 | 1574514 | 1575113 | 600 | - | PROTEIN:peptidase C39, bacteriocin processing |
| Dgeo_1491 | Dgeo_1491 | 1575132 | 1578512 | 3381 | - | PROTEIN:SNF2-related |
| Dgeo_1492 | Dgeo_1492 | 1578514 | 1580217 | 1704 | - | PROTEIN:peptide chain release factor 3 |
| Dgeo_1493 | Dgeo_1493 | 1580369 | 1580842 | 474 | + | PROTEIN:putative 2-nitropropane dioxygenase |
| Dgeo_1494 | Dgeo_1494 | 1580820 | 1581386 | 567 | + | PROTEIN:2-nitropropane dioxygenase, NPD |
| Dgeo_1495 | Dgeo_1495 | 1581374 | 1582495 | 1122 | + | PROTEIN:peptidase S1 and S6, chymotrypsin/Hap |
| Dgeo_1496 | Dgeo_1496 | 1582501 | 1583094 | 594 | + | PROTEIN:LuxR family transcriptional regulator |
| Dgeo_1497 | Dgeo_1497 | 1583180 | 1583965 | 786 | + | PROTEIN:purine nucleoside phosphorylase |
| Dgeo_1498 | Dgeo_1498 | 1583998 | 1585281 | 1284 | - | PROTEIN:NusB/RsmB/TIM44 |
| Dgeo_1499 | Dgeo_1499 | 1585441 | 1586691 | 1251 | - | PROTEIN:major facilitator transporter |
| Dgeo_1500 | Dgeo_1500 | 1587019 | 1588200 | 1182 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_1501 | Dgeo_1501 | 1588348 | 1589772 | 1425 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1502 | Dgeo_1502 | 1589769 | 1591145 | 1377 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1503 | Dgeo_1503 | 1591350 | 1593017 | 1668 | - | PROTEIN:hypothetical protein |
| Dgeo_1504 | Dgeo_1504 | 1593014 | 1595164 | 2151 | - | PROTEIN:ATPase |
| Dgeo_1505 | Dgeo_1505 | 1595615 | 1598767 | 3153 | - | PROTEIN:hypothetical protein |
| Dgeo_R0031 | Dgeo_R0031 | 1599558 | 1599632 | 75 | - | RNA:Val tRNA |
| Dgeo_R0032 | Dgeo_R0032 | 1599646 | 1599719 | 74 | - | RNA:Cys tRNA |
| Dgeo_1506 | Dgeo_1506 | 1599843 | 1600712 | 870 | + | PROTEIN:glycerophosphoryl diester phosphodiesteras |
| Dgeo_1507 | Dgeo_1507 | 1600759 | 1602708 | 1950 | - | PROTEIN:threonyl-tRNA synthetase |
| Dgeo_1508 | Dgeo_1508 | 1603024 | 1603269 | 246 | - | PROTEIN:glutaredoxin-like protein protein, YruB |
| Dgeo_1509 | Dgeo_1509 | 1603384 | 1603998 | 615 | - | PROTEIN:translation initiation factor IF-3 |
| Dgeo_1510 | Dgeo_1510 | 1604235 | 1604912 | 678 | + | PROTEIN:NUDIX hydrolase |
| Dgeo_1511 | Dgeo_1511 | 1605019 | 1605723 | 705 | + | PROTEIN:hypothetical protein |
| Dgeo_1512 | Dgeo_1512 | 1605726 | 1606241 | 516 | + | PROTEIN:hypothetical protein |
| Dgeo_1513 | Dgeo_1513 | 1606288 | 1606956 | 669 | + | PROTEIN:recombination protein RecR |
| Dgeo_1514 | Dgeo_1514 | 1607038 | 1607658 | 621 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_1515 | Dgeo_1515 | 1607745 | 1608230 | 486 | + | PROTEIN:hypothetical protein |
| Dgeo_1516 | Dgeo_1516 | 1608271 | 1609374 | 1104 | - | PROTEIN:peptidase M23B |
| Dgeo_1517 | Dgeo_1517 | 1609418 | 1609864 | 447 | - | PROTEIN:D-tyrosyl-tRNA deacylase |
| Dgeo_1518 | Dgeo_1518 | 1609866 | 1610216 | 351 | - | PROTEIN:hypothetical protein |
| Dgeo_1519 | Dgeo_1519 | 1610247 | 1610822 | 576 | - | PROTEIN:ECF subfamily RNA polymerase sigma-24 fact |
| Dgeo_1520 | Dgeo_1520 | 1610952 | 1611299 | 348 | + | PROTEIN:FUN14 |
| Dgeo_1521 | Dgeo_1521 | 1611340 | 1612653 | 1314 | - | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_1522 | Dgeo_1522 | 1612650 | 1613333 | 684 | - | PROTEIN:two component transcriptional regulator |
| Dgeo_1523 | Dgeo_1523 | 1613504 | 1614922 | 1419 | + | PROTEIN:outer membrane efflux protein |
| Dgeo_1524 | Dgeo_1524 | 1614919 | 1615962 | 1044 | + | PROTEIN:hypothetical protein |
| Dgeo_1525 | Dgeo_1525 | 1616058 | 1616642 | 585 | - | PROTEIN:peptidase M22, glycoprotease |
| Dgeo_1526 | Dgeo_1526 | 1616708 | 1617880 | 1173 | + | PROTEIN:2-methylcitrate synthase/citrate synthase |
| Dgeo_1527 | Dgeo_1527 | 1617974 | 1618471 | 498 | - | PROTEIN:hypothetical protein |
| Dgeo_1528 | Dgeo_1528 | 1618499 | 1619494 | 996 | - | PROTEIN:beta-lactamase-like protein |
| Dgeo_1529 | Dgeo_1529 | 1619526 | 1619990 | 465 | - | PROTEIN:hypothetical protein |
| Dgeo_1530 | Dgeo_1530 | 1620084 | 1620650 | 567 | - | PROTEIN:hypothetical protein |
| Dgeo_1531 | Dgeo_1531 | 1620726 | 1622366 | 1641 | + | PROTEIN:Pyrrolo-quinoline quinone |
| Dgeo_1532 | Dgeo_1532 | 1622547 | 1623062 | 516 | + | PROTEIN:hypothetical protein |
| Dgeo_1533 | Dgeo_1533 | 1623109 | 1623750 | 642 | + | PROTEIN:putative cyclase |
| Dgeo_1534 | Dgeo_1534 | 1623747 | 1624928 | 1182 | + | PROTEIN:kynureninase |
| Dgeo_1535 | Dgeo_1535 | 1624991 | 1625455 | 465 | - | PROTEIN:thioesterase superfamily protein |
| Dgeo_1536 | Dgeo_1536 | 1625452 | 1625808 | 357 | - | PROTEIN:transcriptional coactivator/pterin dehydra |
| Dgeo_1537 | Dgeo_1537 | 1626128 | 1628770 | 2643 | + | PROTEIN:DNA mismatch repair protein |
| Dgeo_1538 | Dgeo_1538 | 1628795 | 1630444 | 1650 | + | PROTEIN:DNA mismatch repair protein MutL |
| Dgeo_1539 | Dgeo_1539 | 1630527 | 1631969 | 1443 | + | PROTEIN:glutamyl-tRNA synthetase |
| Dgeo_1540 | Dgeo_1540 | 1631973 | 1633103 | 1131 | - | PROTEIN:WD-40 repeat-containing protein |
| Dgeo_1541 | Dgeo_1541 | 1633175 | 1633630 | 456 | + | PROTEIN:methylmalonyl-CoA mutase, alpha subunit |
| Dgeo_1542 | Dgeo_1542 | 1633623 | 1634810 | 1188 | + | PROTEIN:major facilitator transporter |
| Dgeo_1543 | Dgeo_1543 | 1635191 | 1640326 | 5136 | + | PROTEIN:DEAD/DEAH box helicase-like protein |
| Dgeo_1544 | Dgeo_1544 | 1640323 | 1643124 | 2802 | + | PROTEIN:helicase-like protein |
| Dgeo_1545 | Dgeo_1545 | 1643121 | 1647077 | 3957 | + | PROTEIN:putative type II DNA modification enzyme |
| Dgeo_R0033 | Dgeo_R0033 | 1647172 | 1647261 | 90 | - | RNA:Ser tRNA |
| Dgeo_1546 | Dgeo_1546 | 1647302 | 1647874 | 573 | - | PROTEIN:formyl transferase-like protein |
| Dgeo_1547 | Dgeo_1547 | 1647945 | 1649312 | 1368 | - | PROTEIN:peptidase M20 |
| Dgeo_1548 | Dgeo_1548 | 1649319 | 1650188 | 870 | + | PROTEIN:MerR family transcriptional regulator |
| Dgeo_1549 | Dgeo_1549 | 1650238 | 1651101 | 864 | + | PROTEIN:metallophosphoesterase |
| Dgeo_1550 | Dgeo_1550 | 1651098 | 1651628 | 531 | + | PROTEIN:adenine phosphoribosyltransferase |
| Dgeo_1551 | Dgeo_1551 | 1651662 | 1652162 | 501 | - | PROTEIN:hypothetical protein |
| Dgeo_1552 | Dgeo_1552 | 1652223 | 1653200 | 978 | + | PROTEIN:hypothetical protein |
| Dgeo_1553 | Dgeo_1553 | 1653197 | 1653916 | 720 | + | PROTEIN:hypothetical protein |
| Dgeo_1554 | Dgeo_1554 | 1654046 | 1654672 | 627 | - | PROTEIN:NLP/P60 |
| Dgeo_1555 | Dgeo_1555 | 1655025 | 1655441 | 417 | + | PROTEIN:hypothetical protein |
| Dgeo_1556 | Dgeo_1556 | 1655497 | 1656189 | 693 | + | PROTEIN:phage SPO1 DNA polymerase-related protein |
| Dgeo_1557 | Dgeo_1557 | 1656263 | 1657258 | 996 | + | PROTEIN:Allergen V5/Tpx-1 related |
| Dgeo_1558 | Dgeo_1558 | 1657272 | 1658519 | 1248 | - | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_1559 | Dgeo_1559 | 1658516 | 1659010 | 495 | - | PROTEIN:hypothetical protein |
| Dgeo_1560 | Dgeo_1560 | 1659226 | 1660137 | 912 | + | PROTEIN:S-layer-like protein array-related protein |
| Dgeo_1561 | Dgeo_1561 | 1661054 | 1662139 | 1086 | + | PROTEIN:pyruvate dehydrogenase (lipoamide) |
| Dgeo_1562 | Dgeo_1562 | 1662136 | 1663140 | 1005 | + | PROTEIN:transketolase, central region |
| Dgeo_1563 | Dgeo_1563 | 1663199 | 1664464 | 1266 | + | PROTEIN:Na+/H+ antiporter NhaA |
| Dgeo_1564 | Dgeo_1564 | 1664722 | 1665720 | 999 | + | PROTEIN:hypothetical protein |
| Dgeo_1565 | Dgeo_1565 | 1665833 | 1667413 | 1581 | - | PROTEIN:60 kDa inner membrane insertion protein |
| Dgeo_1566 | Dgeo_1566 | 1667410 | 1667691 | 282 | - | PROTEIN:hypothetical protein |
| Dgeo_1567 | Dgeo_1567 | 1667688 | 1668119 | 432 | - | PROTEIN:ribonuclease P protein component |
| Dgeo_1568 | rpmH | 1668209 | 1668352 | 144 | - | PROTEIN:50S ribosomal protein L34 |
| Dgeo_1569 | Dgeo_1569 | 1668654 | 1669703 | 1050 | + | PROTEIN:hypothetical protein |
| Dgeo_1570 | Dgeo_1570 | 1669704 | 1670363 | 660 | + | PROTEIN:putative transcriptional regulator |
| Dgeo_1571 | Dgeo_1571 | 1670363 | 1670971 | 609 | + | PROTEIN:hypothetical protein |
| Dgeo_1572 | Dgeo_1572 | 1671359 | 1672498 | 1140 | - | PROTEIN:bifunctional 3-deoxy-7-phosphoheptulonate |
| Dgeo_1573 | Dgeo_1573 | 1672491 | 1672925 | 435 | - | PROTEIN:globin |
| Dgeo_1574 | Dgeo_1574 | 1673419 | 1674516 | 1098 | + | PROTEIN:translation-associated GTPase |
| Dgeo_1575 | Dgeo_1575 | 1674629 | 1675141 | 513 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_1576 | Dgeo_1576 | 1675211 | 1676194 | 984 | - | PROTEIN:thioredoxin reductase |
| Dgeo_R0034 | Dgeo_R0034 | 1676412 | 1676485 | 74 | - | RNA:Gln tRNA |
| Dgeo_1577 | rpsA | 1676893 | 1678656 | 1764 | + | PROTEIN:30S ribosomal protein S1 |
| Dgeo_1578 | Dgeo_1578 | 1678744 | 1679727 | 984 | + | PROTEIN:oxidoreductase-like protein |
| Dgeo_1579 | Dgeo_1579 | 1679732 | 1682383 | 2652 | - | PROTEIN:DNA internalization-related competence pro |
| Dgeo_1580 | Dgeo_1580 | 1682398 | 1683309 | 912 | + | PROTEIN:hypothetical protein |
| Dgeo_1581 | Dgeo_1581 | 1683430 | 1683729 | 300 | + | PROTEIN:hypothetical protein |
| Dgeo_1582 | Dgeo_1582 | 1683779 | 1684852 | 1074 | - | PROTEIN:putative RNA methylase |
| Dgeo_1583 | Dgeo_1583 | 1684952 | 1686328 | 1377 | + | PROTEIN:K+ transporter Trk |
| Dgeo_1584 | Dgeo_1584 | 1686401 | 1687060 | 660 | + | PROTEIN:TrkA-N |
| Dgeo_1585 | Dgeo_1585 | 1687138 | 1687596 | 459 | - | PROTEIN:hypothetical protein |
| Dgeo_1586 | Dgeo_1586 | 1687626 | 1688429 | 804 | - | PROTEIN:beta-lactamase-like protein |
| Dgeo_1587 | Dgeo_1587 | 1688426 | 1689214 | 789 | - | PROTEIN:indole-3-glycerol-phosphate synthase |
| Dgeo_1588 | Dgeo_1588 | 1689294 | 1689821 | 528 | + | PROTEIN:hypoxanthine phosphoribosyltransferase |
| Dgeo_1589 | Dgeo_1589 | 1689868 | 1690137 | 270 | + | PROTEIN:Na+/H+ antiporter NhaD and related arsenit |
| Dgeo_1590 | Dgeo_1590 | 1690409 | 1691128 | 720 | + | PROTEIN:arsenical pump membrane protein |
| Dgeo_1591 | Dgeo_1591 | 1691138 | 1691560 | 423 | + | PROTEIN:hypothetical protein |
| Dgeo_1592 | Dgeo_1592 | 1691689 | 1692147 | 459 | + | PROTEIN:hypothetical protein |
| Dgeo_1593 | Dgeo_1593 | 1692175 | 1693872 | 1698 | - | PROTEIN:VanW |
| Dgeo_1594 | Dgeo_1594 | 1693945 | 1695027 | 1083 | - | PROTEIN:hypothetical protein |
| Dgeo_1595 | Dgeo_1595 | 1695041 | 1695838 | 798 | - | PROTEIN:enoyl-CoA hydratase/isomerase |
| Dgeo_1596 | Dgeo_1596 | 1695899 | 1696615 | 717 | - | PROTEIN:purine phosphorylases family protein 1 |
| Dgeo_1597 | Dgeo_1597 | 1696963 | 1698213 | 1251 | + | PROTEIN:S-layer-like protein region |
| Dgeo_1598 | Dgeo_1598 | 1698345 | 1698677 | 333 | + | PROTEIN:hypothetical protein |
| Dgeo_1599 | Dgeo_1599 | 1698674 | 1700782 | 2109 | + | PROTEIN:single-stranded-DNA-specific exonuclease R |
| Dgeo_1600 | Dgeo_1600 | 1700863 | 1701888 | 1026 | - | PROTEIN:hypothetical protein |
| Dgeo_R0035 | Dgeo_R0035 | 1702114 | 1702190 | 77 | - | RNA:Arg tRNA |
| Dgeo_1601 | Dgeo_1601 | 1702246 | 1704153 | 1908 | + | PROTEIN:PpiC-type peptidyl-prolyl cis-trans isomer |
| Dgeo_1602 | Dgeo_1602 | 1704271 | 1705272 | 1002 | + | PROTEIN:tryptophanyl-tRNA synthetase |
| Dgeo_1603 | Dgeo_1603 | 1705273 | 1706004 | 732 | + | PROTEIN:hypothetical protein |
| Dgeo_1604 | Dgeo_1604 | 1706008 | 1706973 | 966 | - | PROTEIN:N-acetyl-gamma-glutamyl-phosphate reductas |
| Dgeo_1605 | Dgeo_1605 | 1706986 | 1707909 | 924 | - | PROTEIN:ornithine carbamoyltransferase |
| Dgeo_1606 | Dgeo_1606 | 1708127 | 1709023 | 897 | + | PROTEIN:hypothetical protein |
| Dgeo_1607 | Dgeo_1607 | 1709033 | 1710028 | 996 | + | PROTEIN:ribonuclease Z |
| Dgeo_1608 | Dgeo_1608 | 1710044 | 1710835 | 792 | + | PROTEIN:histidinol-phosphatase |
| Dgeo_1609 | Dgeo_1609 | 1710828 | 1712546 | 1719 | + | PROTEIN:PHP-like protein |
| Dgeo_1610 | Dgeo_1610 | 1712606 | 1713772 | 1167 | - | PROTEIN:major facilitator transporter |
| Dgeo_1611 | Dgeo_1611 | 1713887 | 1714387 | 501 | + | PROTEIN:hypothetical protein |
| Dgeo_1612 | Dgeo_1612 | 1714384 | 1715061 | 678 | - | PROTEIN:isoprenylcysteine carboxyl methyltransfera |
| Dgeo_1613 | Dgeo_1613 | 1715191 | 1716129 | 939 | - | PROTEIN:LAO/AO transport system ATPase |
| Dgeo_1614 | Dgeo_1614 | 1716136 | 1717731 | 1596 | - | PROTEIN:diguanylate cyclase with PAS/PAC sensor |
| Dgeo_1615 | Dgeo_1615 | 1717895 | 1718608 | 714 | - | PROTEIN:phosphoglycerate mutase |
| Dgeo_1616 | Dgeo_1616 | 1718605 | 1719672 | 1068 | - | PROTEIN:phosphoribosylaminoimidazole synthetase |
| Dgeo_1617 | Dgeo_1617 | 1719718 | 1720422 | 705 | - | PROTEIN:DedA-like protein |
| Dgeo_1618 | Dgeo_1618 | 1720492 | 1721481 | 990 | + | PROTEIN:polyprenyl synthetase |
| Dgeo_1619 | Dgeo_1619 | 1721507 | 1722295 | 789 | - | PROTEIN:N-formylglutamate amidohydrolase |
| Dgeo_1620 | Dgeo_1620 | 1722396 | 1723472 | 1077 | + | PROTEIN:DNA replication and repair protein RecF |
| Dgeo_1621 | Dgeo_1621 | 1723527 | 1724390 | 864 | + | PROTEIN:hypothetical protein |
| Dgeo_1622 | Dgeo_1622 | 1724394 | 1725617 | 1224 | - | PROTEIN:rRNA (guanine-N(2)-)-methyltransferase |
| Dgeo_1623 | Dgeo_1623 | 1725635 | 1726309 | 675 | + | PROTEIN:ribonuclease H |
| Dgeo_1624 | Dgeo_1624 | 1726772 | 1728559 | 1788 | + | PROTEIN:GTP-binding protein LepA |
| Dgeo_1625 | Dgeo_1625 | 1728792 | 1729865 | 1074 | + | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_1626 | Dgeo_1626 | 1729862 | 1730446 | 585 | + | PROTEIN:two component LuxR family transcriptional |
| Dgeo_1627 | Dgeo_1627 | 1730597 | 1731778 | 1182 | + | PROTEIN:aminotransferase, class I and II |
| Dgeo_1628 | Dgeo_1628 | 1731855 | 1732478 | 624 | - | PROTEIN:nitroreductase |
| Dgeo_1629 | Dgeo_1629 | 1732554 | 1733309 | 756 | + | PROTEIN:hypothetical protein |
| Dgeo_1630 | Dgeo_1630 | 1733429 | 1734514 | 1086 | + | PROTEIN:UDP-N-acetylglucosamine--N-acetylmuramyl-( |
| Dgeo_1631 | Dgeo_1631 | 1734544 | 1735941 | 1398 | + | PROTEIN:UDP-N-acetylmuramate--alanine ligase |
| Dgeo_1632 | Dgeo_1632 | 1735938 | 1736825 | 888 | + | PROTEIN:UDP-N-acetylenolpyruvoylglucosamine reduct |
| Dgeo_1633 | Dgeo_1633 | 1736904 | 1737653 | 750 | + | PROTEIN:cell division protein FtsQ |
| Dgeo_1634 | Dgeo_1634 | 1737650 | 1738996 | 1347 | + | PROTEIN:cell division protein FtsA |
| Dgeo_1635 | Dgeo_1635 | 1739122 | 1740207 | 1086 | + | PROTEIN:cell division protein FtsZ |
| Dgeo_1636 | Dgeo_1636 | 1740541 | 1741695 | 1155 | + | PROTEIN:extracellular ligand-binding receptor |
| Dgeo_1637 | Dgeo_1637 | 1741762 | 1742772 | 1011 | - | PROTEIN:6-phosphofructokinase |
| Dgeo_1638 | Dgeo_1638 | 1742802 | 1743644 | 843 | - | PROTEIN:hypothetical protein |
| Dgeo_1639 | Dgeo_1639 | 1743729 | 1743962 | 234 | - | PROTEIN:hypothetical protein |
| Dgeo_1640 | Dgeo_1640 | 1744030 | 1744458 | 429 | - | PROTEIN:hypothetical protein |
| Dgeo_1641 | Dgeo_1641 | 1744560 | 1745006 | 447 | + | PROTEIN:hypothetical protein |
| Dgeo_1642 | Dgeo_1642 | 1745080 | 1745526 | 447 | - | PROTEIN:hypothetical protein |
| Dgeo_1643 | Dgeo_1643 | 1745580 | 1748498 | 2919 | - | PROTEIN:peptidase M16C associated |
| Dgeo_1644 | Dgeo_1644 | 1748593 | 1749528 | 936 | + | PROTEIN:putative Ig |
| Dgeo_1645 | Dgeo_1645 | 1749525 | 1750367 | 843 | + | PROTEIN:hypothetical protein |
| Dgeo_1646 | Dgeo_1646 | 1750364 | 1750900 | 537 | + | PROTEIN:hypothetical protein |
| Dgeo_1647 | Dgeo_1647 | 1750953 | 1751153 | 201 | + | PROTEIN:RNA-binding S4 |
| Dgeo_1648 | Dgeo_1648 | 1751150 | 1751692 | 543 | + | PROTEIN:hypothetical protein |
| Dgeo_1649 | Dgeo_1649 | 1751574 | 1752227 | 654 | - | PROTEIN:peptidase S26A, signal peptidase I |
| Dgeo_1650 | Dgeo_1650 | 1752301 | 1752723 | 423 | + | PROTEIN:peptidylprolyl isomerase |
| Dgeo_1651 | Dgeo_1651 | 1752817 | 1753068 | 252 | - | PROTEIN:hypothetical protein |
| Dgeo_1652 | Dgeo_1652 | 1753096 | 1754046 | 951 | + | PROTEIN:purine nucleosidase |
| Dgeo_1653 | Dgeo_1653 | 1754083 | 1754502 | 420 | - | PROTEIN:Mov34/MPN/PAD-1 |
| Dgeo_1654 | Dgeo_1654 | 1754564 | 1757704 | 3141 | + | PROTEIN:cell divisionFtsK/SpoIIIE |
| Dgeo_1655 | Dgeo_1655 | 1757921 | 1759711 | 1791 | + | PROTEIN:beta-Ig-H3/fasciclin |
| Dgeo_1656 | Dgeo_1656 | 1759798 | 1760634 | 837 | - | PROTEIN:metallophosphoesterase |
| Dgeo_1657 | Dgeo_1657 | 1760693 | 1761661 | 969 | + | PROTEIN:cysteine synthase A |
| Dgeo_1658 | Dgeo_1658 | 1761677 | 1762378 | 702 | - | PROTEIN:hypothetical protein |
| Dgeo_1659 | Dgeo_1659 | 1762510 | 1763799 | 1290 | - | PROTEIN:polysaccharide deacetylase |
| Dgeo_1660 | Dgeo_1660 | 1763867 | 1764496 | 630 | + | PROTEIN:DNA-3-methyladenine glycosylase |
| Dgeo_1661 | Dgeo_1661 | 1764524 | 1765672 | 1149 | + | PROTEIN:hypothetical protein |
| Dgeo_1662 | Dgeo_1662 | 1765669 | 1766667 | 999 | - | PROTEIN:aminoglycoside phosphotransferase |
| Dgeo_1664 | Dgeo_1664 | 1767019 | 1768353 | 1335 | + | PROTEIN:phosphoglucosamine mutase |
| Dgeo_1665 | Dgeo_1665 | 1768756 | 1770144 | 1389 | + | PROTEIN:Ig-like protein, group 2 |
| Dgeo_1666 | Dgeo_1666 | 1770264 | 1772987 | 2724 | + | PROTEIN:DNA polymerase I |
| Dgeo_1667 | Dgeo_1667 | 1773028 | 1773627 | 600 | + | PROTEIN:HAD family hydrolase |
| Dgeo_1668 | Dgeo_1668 | 1773629 | 1774651 | 1023 | - | PROTEIN:hypothetical protein |
| Dgeo_1669 | Dgeo_1669 | 1774866 | 1775258 | 393 | - | PROTEIN:putative aldehyde reductase |
| Dgeo_1670 | Dgeo_1670 | 1775468 | 1775797 | 330 | - | PROTEIN:hypothetical protein |
| Dgeo_1671 | Dgeo_1671 | 1776053 | 1776556 | 504 | - | PROTEIN:hypothetical protein |
| Dgeo_1672 | Dgeo_1672 | 1777087 | 1778232 | 1146 | + | PROTEIN:putative PAS/PAC sensor protein |
| Dgeo_1673 | Dgeo_1673 | 1778499 | 1778945 | 447 | + | PROTEIN:IS1 transposase |
| Dgeo_1674 | Dgeo_1674 | 1778942 | 1779904 | 963 | + | PROTEIN:diguanylate cyclase |
| Dgeo_1675 | Dgeo_1675 | 1780254 | 1780994 | 741 | + | PROTEIN:ABC transporter related |
| Dgeo_1676 | Dgeo_1676 | 1780984 | 1782225 | 1242 | + | PROTEIN:ABC-2 type transporter |
| Dgeo_1677 | Dgeo_1677 | 1782332 | 1784023 | 1692 | - | PROTEIN:PRC-barrel domain-containing protein |
| Dgeo_1678 | Dgeo_1678 | 1784383 | 1785153 | 771 | + | PROTEIN:hypothetical protein |
| Dgeo_1679 | Dgeo_1679 | 1785150 | 1786238 | 1089 | + | PROTEIN:mandelate racemase/muconate lactonizing-li |
| Dgeo_1680 | Dgeo_1680 | 1786310 | 1786585 | 276 | + | PROTEIN:hypothetical protein |
| Dgeo_1681 | Dgeo_1681 | 1786663 | 1787997 | 1335 | - | PROTEIN:peptidase S8 and S53, subtilisin, kexin, s |
| Dgeo_1682 | Dgeo_1682 | 1788130 | 1790850 | 2721 | - | PROTEIN:aconitate hydratase |
| Dgeo_1683 | Dgeo_1683 | 1791188 | 1791523 | 336 | + | PROTEIN:hypothetical protein |
| Dgeo_1684 | Dgeo_1684 | 1791546 | 1792682 | 1137 | - | PROTEIN:chalcone and stilbene synthases-like prote |
| Dgeo_1685 | Dgeo_1685 | 1792737 | 1793639 | 903 | - | PROTEIN:aldo/keto reductase |
| Dgeo_1686 | Dgeo_1686 | 1793708 | 1794538 | 831 | - | PROTEIN:nitroreductase |
| Dgeo_1687 | Dgeo_1687 | 1794561 | 1794887 | 327 | - | PROTEIN:hypothetical protein |
| Dgeo_1688 | Dgeo_1688 | 1794942 | 1795382 | 441 | + | PROTEIN:hypothetical protein |
| Dgeo_1689 | Dgeo_1689 | 1795493 | 1795942 | 450 | - | PROTEIN:nucleoside-diphosphate kinase |
| Dgeo_1690 | Dgeo_1690 | 1796125 | 1798122 | 1998 | + | PROTEIN:inner-membrane translocator |
| Dgeo_1691 | Dgeo_1691 | 1798123 | 1799037 | 915 | + | PROTEIN:inner-membrane translocator |
| Dgeo_1692 | Dgeo_1692 | 1799106 | 1800095 | 990 | - | PROTEIN:LysR family transcriptional regulator |
| Dgeo_1693 | Dgeo_1693 | 1800225 | 1800545 | 321 | + | PROTEIN:hypothetical protein |
| Dgeo_1694 | Dgeo_1694 | 1800605 | 1801939 | 1335 | + | PROTEIN:peptidoglycan-binding LysM |
| Dgeo_1695 | Dgeo_1695 | 1802078 | 1802368 | 291 | + | PROTEIN:ribosome-binding factor A |
| Dgeo_1696 | Dgeo_1696 | 1802365 | 1802781 | 417 | + | PROTEIN:4-hydroxybenzoyl-CoA thioesterase |
| Dgeo_1697 | Dgeo_1697 | 1802840 | 1803316 | 477 | + | PROTEIN:NUDIX hydrolase |
| Dgeo_1698 | Dgeo_1698 | 1803326 | 1804537 | 1212 | + | PROTEIN:peptidase M1, membrane alanine aminopeptid |
| Dgeo_1699 | Dgeo_1699 | 1804578 | 1805375 | 798 | - | PROTEIN:transposase, IS4 |
| Dgeo_1700 | Dgeo_1700 | 1805639 | 1806163 | 525 | - | PROTEIN:hypothetical protein |
| Dgeo_1701 | Dgeo_1701 | 1806215 | 1807345 | 1131 | - | PROTEIN:hypothetical protein |
| Dgeo_1702 | Dgeo_1702 | 1807758 | 1808849 | 1092 | + | PROTEIN:transposase, IS4 |
| Dgeo_1703 | Dgeo_1703 | 1808857 | 1809714 | 858 | - | PROTEIN:phage integrase |
| Dgeo_1704 | Dgeo_1704 | 1809837 | 1812857 | 3021 | + | PROTEIN:transposase Tn3 |
| Dgeo_1705 | Dgeo_1705 | 1812864 | 1813175 | 312 | - | PROTEIN:hypothetical protein |
| Dgeo_1706 | Dgeo_1706 | 1813191 | 1814885 | 1695 | + | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_1707 | Dgeo_1707 | 1814882 | 1815937 | 1056 | + | PROTEIN:hypothetical protein |
| Dgeo_1708 | Dgeo_1708 | 1815934 | 1816599 | 666 | + | PROTEIN:two component LuxR family transcriptional |
| Dgeo_1709 | Dgeo_1709 | 1816747 | 1817238 | 492 | + | PROTEIN:cupin 2, barrel |
| Dgeo_1710 | Dgeo_1710 | 1817324 | 1818070 | 747 | - | PROTEIN:chlorite dismutase |
| Dgeo_1711 | Dgeo_1711 | 1818154 | 1819176 | 1023 | - | PROTEIN:aldo/keto reductase |
| Dgeo_1712 | Dgeo_1712 | 1819273 | 1819695 | 423 | + | PROTEIN:glyoxalase/bleomycin resistance protein/di |
| Dgeo_1713 | Dgeo_1713 | 1819702 | 1820817 | 1116 | - | PROTEIN:hypothetical protein |
| Dgeo_1714 | cysS | 1821063 | 1822556 | 1494 | + | PROTEIN:cysteinyl-tRNA synthetase |
| Dgeo_1715 | Dgeo_1715 | 1822755 | 1823333 | 579 | + | PROTEIN:hypothetical protein |
| Dgeo_1716 | Dgeo_1716 | 1824040 | 1825554 | 1515 | + | PROTEIN:glycyl-tRNA synthetase |
| Dgeo_1717 | Dgeo_1717 | 1825656 | 1826387 | 732 | - | PROTEIN:hypothetical protein |
| Dgeo_1718 | Dgeo_1718 | 1826457 | 1827044 | 588 | - | PROTEIN:uracil-DNA glycosylase superfamily protein |
| Dgeo_1719 | Dgeo_1719 | 1827037 | 1827633 | 597 | - | PROTEIN:oxidoreductase, molybdopterin binding |
| Dgeo_1720 | Dgeo_1720 | 1827861 | 1828790 | 930 | + | PROTEIN:hypothetical protein |
| Dgeo_1721 | Dgeo_1721 | 1829016 | 1829348 | 333 | + | PROTEIN:hypothetical protein |
| Dgeo_1722 | Dgeo_1722 | 1829341 | 1830957 | 1617 | + | PROTEIN:Dak phosphatase |
| Dgeo_1723 | Dgeo_1723 | 1831035 | 1832324 | 1290 | + | PROTEIN:UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6- |
| Dgeo_1724 | Dgeo_1724 | 1832383 | 1833159 | 777 | + | PROTEIN:hypothetical protein |
| Dgeo_1725 | Dgeo_1725 | 1833429 | 1834607 | 1179 | + | PROTEIN:type IV pilus assembly protein PilM |
| Dgeo_1726 | Dgeo_1726 | 1834600 | 1835325 | 726 | + | PROTEIN:fimbrial assembly |
| Dgeo_1727 | Dgeo_1727 | 1835315 | 1835947 | 633 | + | PROTEIN:type IV pilus assembly protein PilO |
| Dgeo_1728 | Dgeo_1728 | 1835944 | 1837137 | 1194 | + | PROTEIN:hypothetical protein |
| Dgeo_1729 | Dgeo_1729 | 1837141 | 1839162 | 2022 | + | PROTEIN:type II and III secretion system protein |
| Dgeo_1730 | Dgeo_1730 | 1839278 | 1840426 | 1149 | + | PROTEIN:chorismate synthase |
| Dgeo_1731 | Dgeo_1731 | 1840599 | 1841141 | 543 | + | PROTEIN:shikimate kinase |
| Dgeo_1732 | Dgeo_1732 | 1841128 | 1842234 | 1107 | + | PROTEIN:3-dehydroquinate synthase |
| Dgeo_1733 | Dgeo_1733 | 1842281 | 1842766 | 486 | + | PROTEIN:3-dehydroquinate dehydratase |
| Dgeo_1734 | rplM | 1842937 | 1843380 | 444 | + | PROTEIN:50S ribosomal protein L13 |
| Dgeo_1735 | rpsI | 1843383 | 1843778 | 396 | + | PROTEIN:30S ribosomal protein S9 |
| Dgeo_1736 | Dgeo_1736 | 1843860 | 1845311 | 1452 | + | PROTEIN:ABC transporter related |
| Dgeo_1737 | Dgeo_1737 | 1845308 | 1846153 | 846 | - | PROTEIN:HAD family hydrolase |
| Dgeo_1738 | rplS | 1846184 | 1846660 | 477 | - | PROTEIN:50S ribosomal protein L19 |
| Dgeo_1739 | Dgeo_1739 | 1846970 | 1847740 | 771 | + | PROTEIN:hypothetical protein |
| Dgeo_1740 | Dgeo_1740 | 1847719 | 1848270 | 552 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_1741 | Dgeo_1741 | 1848458 | 1849174 | 717 | + | PROTEIN:lipoate-protein ligase B |
| Dgeo_1742 | Dgeo_1742 | 1849179 | 1850165 | 987 | + | PROTEIN:lipoyl synthase |
| Dgeo_1743 | Dgeo_1743 | 1850202 | 1850711 | 510 | + | PROTEIN:short chain dehydrogenase |
| Dgeo_R0036 | Dgeo_R0036 | 1850752 | 1850828 | 77 | - | RNA:Ile tRNA |
| Dgeo_1744 | Dgeo_1744 | 1850978 | 1852138 | 1161 | + | PROTEIN:diguanylate cyclase |
| Dgeo_1745 | Dgeo_1745 | 1852145 | 1852429 | 285 | - | PROTEIN:hypothetical protein |
| Dgeo_1746 | Dgeo_1746 | 1852512 | 1855577 | 3066 | + | PROTEIN:alpha amylase, catalytic region |
| Dgeo_1747 | Dgeo_1747 | 1855691 | 1856329 | 639 | + | PROTEIN:hypothetical protein |
| Dgeo_1748 | Dgeo_1748 | 1856348 | 1857346 | 999 | + | PROTEIN:phosphate transporter |
| Dgeo_1749 | Dgeo_1749 | 1857419 | 1858219 | 801 | - | PROTEIN:pantothenate kinase |
| Dgeo_1750 | Dgeo_1750 | 1858317 | 1860227 | 1911 | + | PROTEIN:AMP-dependent synthetase and ligase |
| Dgeo_1751 | Dgeo_1751 | 1860284 | 1860991 | 708 | - | PROTEIN:hypothetical protein |
| Dgeo_1752 | Dgeo_1752 | 1861049 | 1862959 | 1911 | - | PROTEIN:hypothetical protein |
| Dgeo_1753 | Dgeo_1753 | 1862967 | 1863383 | 417 | - | PROTEIN:biopolymer transport protein ExbD/TolR |
| Dgeo_1754 | Dgeo_1754 | 1863380 | 1864072 | 693 | - | PROTEIN:MotA/TolQ/ExbB proton channel |
| Dgeo_1755 | nfrB | 1864188 | 1866200 | 2013 | - | PROTEIN:bacteriophage N4 adsorption protein B |
| Dgeo_1756 | Dgeo_1756 | 1866190 | 1867266 | 1077 | - | PROTEIN:hypothetical protein |
| Dgeo_1757 | Dgeo_1757 | 1867280 | 1869007 | 1728 | - | PROTEIN:hypothetical protein |
| Dgeo_1758 | Dgeo_1758 | 1869199 | 1870374 | 1176 | + | PROTEIN:aminotransferase, class I and II |
| Dgeo_1759 | Dgeo_1759 | 1870393 | 1872360 | 1968 | - | PROTEIN:acetyl-CoA synthetase |
| Dgeo_1760 | Dgeo_1760 | 1872563 | 1874155 | 1593 | - | PROTEIN:SSS family solute/sodium (Na+) symporter |
| Dgeo_1761 | Dgeo_1761 | 1874152 | 1874451 | 300 | - | PROTEIN:hypothetical protein |
| Dgeo_1762 | Dgeo_1762 | 1874678 | 1875679 | 1002 | + | PROTEIN:molybdenum cofactor biosynthesis protein A |
| Dgeo_1763 | Dgeo_1763 | 1875805 | 1876497 | 693 | + | PROTEIN:GntR family transcriptional regulator |
| Dgeo_1764 | Dgeo_1764 | 1876624 | 1877259 | 636 | + | PROTEIN:TetR family transcriptional regulator |
| Dgeo_1765 | Dgeo_1765 | 1877347 | 1878825 | 1479 | + | PROTEIN:2-phosphoglycerate kinase |
| Dgeo_1766 | Dgeo_1766 | 1878843 | 1880003 | 1161 | - | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_1767 | Dgeo_1767 | 1880005 | 1881357 | 1353 | + | PROTEIN:SpoIID/LytB domain-containing protein |
| Dgeo_1768 | Dgeo_1768 | 1881347 | 1884157 | 2811 | + | PROTEIN:hypothetical protein |
| Dgeo_1769 | Dgeo_1769 | 1884189 | 1885301 | 1113 | + | PROTEIN:hypothetical protein |
| Dgeo_1770 | Dgeo_1770 | 1885311 | 1886327 | 1017 | - | PROTEIN:beta-lactamase |
| Dgeo_R0037 | Dgeo_R0037 | 1886401 | 1886476 | 76 | + | RNA:Asp tRNA |
| Dgeo_1771 | Dgeo_1771 | 1886477 | 1888597 | 2121 | + | PROTEIN:arginine decarboxylase |
| Dgeo_1772 | Dgeo_1772 | 1888987 | 1889406 | 420 | + | PROTEIN:thioesterase superfamily protein |
| Dgeo_1773 | Dgeo_1773 | 1889484 | 1889762 | 279 | + | PROTEIN:NrdI |
| Dgeo_1774 | Dgeo_1774 | 1889891 | 1891840 | 1950 | + | PROTEIN:ribonucleoside-diphosphate reductase, alph |
| Dgeo_1775 | nrdF | 1891939 | 1892898 | 960 | + | PROTEIN:ribonucleotide-diphosphate reductase subun |
| Dgeo_1776 | Dgeo_1776 | 1892906 | 1893151 | 246 | + | PROTEIN:thioredoxin-related protein |
| Dgeo_1777 | Dgeo_1777 | 1893180 | 1894412 | 1233 | + | PROTEIN:(Uracil-5)-methyltransferase |
| Dgeo_1778 | Dgeo_1778 | 1894504 | 1894854 | 351 | + | PROTEIN:hypothetical protein |
| Dgeo_1779 | Dgeo_1779 | 1894875 | 1896137 | 1263 | + | PROTEIN:amine oxidase |
| Dgeo_1780 | Dgeo_1780 | 1896154 | 1896774 | 621 | + | PROTEIN:hypothetical protein |
| Dgeo_1781 | Dgeo_1781 | 1896811 | 1897455 | 645 | - | PROTEIN:rhomboid-like protein protein |
| Dgeo_1782 | Dgeo_1782 | 1897523 | 1898542 | 1020 | - | PROTEIN:aspartate-semialdehyde dehydrogenase, USG- |
| Dgeo_1783 | Dgeo_1783 | 1898693 | 1898992 | 300 | - | PROTEIN:hypothetical protein |
| Dgeo_1784 | Dgeo_1784 | 1899216 | 1900274 | 1059 | - | PROTEIN:transposase, IS4 |
| Dgeo_1785 | Dgeo_1785 | 1900541 | 1901602 | 1062 | + | PROTEIN:hypothetical protein |
| Dgeo_1786 | Dgeo_1786 | 1901722 | 1902264 | 543 | + | PROTEIN:hypothetical protein |
| Dgeo_1787 | Dgeo_1787 | 1902561 | 1903520 | 960 | + | PROTEIN:hypothetical protein |
| Dgeo_1788 | Dgeo_1788 | 1903366 | 1903962 | 597 | - | PROTEIN:hypothetical protein |
| Dgeo_1789 | Dgeo_1789 | 1904065 | 1905528 | 1464 | + | PROTEIN:diguanylate cyclase with GAF sensor |
| Dgeo_1790 | Dgeo_1790 | 1905509 | 1905985 | 477 | + | PROTEIN:glyoxalase/bleomycin resistance protein/di |
| Dgeo_1791 | Dgeo_1791 | 1905982 | 1906356 | 375 | + | PROTEIN:VanZ like protein |
| Dgeo_1792 | Dgeo_1792 | 1906366 | 1907319 | 954 | + | PROTEIN:ATPase |
| Dgeo_1793 | Dgeo_1793 | 1907391 | 1910222 | 2832 | + | PROTEIN:transglutaminase-like protein |
| Dgeo_1794 | Dgeo_1794 | 1910418 | 1911962 | 1545 | + | PROTEIN:threonine dehydratase |
| Dgeo_1795 | Dgeo_1795 | 1912002 | 1912964 | 963 | - | PROTEIN:hypothetical protein |
| Dgeo_1796 | Dgeo_1796 | 1912976 | 1913710 | 735 | - | PROTEIN:twin-arginine translocation pathway signal |
| Dgeo_1797 | Dgeo_1797 | 1913864 | 1914649 | 786 | - | PROTEIN:enoyl-CoA hydratase/isomerase |
| Dgeo_1798 | Dgeo_1798 | 1915042 | 1915809 | 768 | + | PROTEIN:hypothetical protein |
| Dgeo_1799 | Dgeo_1799 | 1915915 | 1916580 | 666 | + | PROTEIN:major intrinsic protein |
| Dgeo_R0038 | Dgeo_R0038 | 1916667 | 1916743 | 77 | - | RNA:Arg tRNA |
| Dgeo_R0039 | Dgeo_R0039 | 1916762 | 1916854 | 93 | - | RNA:Ser tRNA |
| Dgeo_R0040 | Dgeo_R0040 | 1916880 | 1916967 | 88 | - | RNA:Ser tRNA |
| Dgeo_1800 | Dgeo_1800 | 1917036 | 1918298 | 1263 | - | PROTEIN:Nitrilase/cyanide hydratase and apolipopro |
| Dgeo_1801 | Dgeo_1801 | 1918331 | 1918948 | 618 | + | PROTEIN:hypothetical protein |
| Dgeo_1802 | Dgeo_1802 | 1918945 | 1919919 | 975 | + | PROTEIN:allophanate hydrolase subunit 2 |
| Dgeo_1803 | Dgeo_1803 | 1919898 | 1920644 | 747 | + | PROTEIN:LamB/YcsF |
| Dgeo_1804 | Dgeo_1804 | 1920666 | 1921823 | 1158 | - | PROTEIN:deoxyguanosinetriphosphate triphosphohydro |
| Dgeo_1805 | Dgeo_1805 | 1921902 | 1922774 | 873 | - | PROTEIN:ABC-2 type transporter |
| Dgeo_1806 | Dgeo_1806 | 1922771 | 1923655 | 885 | - | PROTEIN:ABC transporter related |
| Dgeo_1807 | Dgeo_1807 | 1923791 | 1924792 | 1002 | + | PROTEIN:transposase, IS4 |
| Dgeo_1808 | Dgeo_1808 | 1924997 | 1925578 | 582 | + | PROTEIN:NUDIX hydrolase |
| Dgeo_1809 | Dgeo_1809 | 1925575 | 1926486 | 912 | + | PROTEIN:riboflavin biosynthesis protein RibF |
| Dgeo_1810 | Dgeo_1810 | 1926565 | 1927404 | 840 | + | PROTEIN:hypothetical protein |
| Dgeo_1811 | Dgeo_1811 | 1927667 | 1928602 | 936 | + | PROTEIN:ABC transporter related |
| Dgeo_1812 | Dgeo_1812 | 1928637 | 1929410 | 774 | + | PROTEIN:putative ABC-2 type transport system perme |
| Dgeo_1813 | Dgeo_1813 | 1929484 | 1929960 | 477 | - | PROTEIN:GreA/GreB family elongation factor |
| Dgeo_1814 | Dgeo_1814 | 1930116 | 1930676 | 561 | + | PROTEIN:hypothetical protein |
| Dgeo_1815 | Dgeo_1815 | 1930743 | 1931333 | 591 | - | PROTEIN:gliding motility protein |
| Dgeo_1816 | Dgeo_1816 | 1931345 | 1931830 | 486 | - | PROTEIN:roadblock/LC7 domain-contain protein |
| Dgeo_1817 | Dgeo_1817 | 1931849 | 1933762 | 1914 | + | PROTEIN:hypothetical protein |
| Dgeo_1818 | Dgeo_1818 | 1933776 | 1934318 | 543 | - | PROTEIN:DNA polymerase III, epsilon subunit |
| Dgeo_1819 | uvsE | 1934355 | 1935290 | 936 | - | PROTEIN:putative UV damage endonuclease |
| Dgeo_1820 | Dgeo_1820 | 1935287 | 1935457 | 171 | - | PROTEIN:hypothetical protein |
| Dgeo_1821 | Dgeo_1821 | 1935573 | 1936640 | 1068 | - | PROTEIN:phospho-2-dehydro-3-deoxyheptonate aldolas |
| Dgeo_1822 | Dgeo_1822 | 1936773 | 1937477 | 705 | + | PROTEIN:hypothetical protein |
| Dgeo_1823 | Dgeo_1823 | 1937729 | 1938049 | 321 | - | PROTEIN:hypothetical protein |
| Dgeo_1824 | Dgeo_1824 | 1938133 | 1938837 | 705 | - | PROTEIN:hypothetical protein |
| Dgeo_1825 | era | 1938852 | 1939781 | 930 | + | PROTEIN:GTP-binding protein Era |
| Dgeo_1826 | Dgeo_1826 | 1939933 | 1940688 | 756 | + | PROTEIN:cytochrome c, class I |
| Dgeo_1827 | Dgeo_1827 | 1940815 | 1942395 | 1581 | + | PROTEIN:twin-arginine translocation pathway signal |
| Dgeo_1828 | Dgeo_1828 | 1942473 | 1942889 | 417 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_1829 | Dgeo_1829 | 1942889 | 1943401 | 513 | - | PROTEIN:metal-dependent hydrolase |
| Dgeo_1830 | Dgeo_1830 | 1943398 | 1943757 | 360 | - | PROTEIN:MazG nucleotide pyrophosphohydrolase |
| Dgeo_1831 | Dgeo_1831 | 1943787 | 1944275 | 489 | - | PROTEIN:hypothetical protein |
| Dgeo_1832 | Dgeo_1832 | 1944414 | 1946285 | 1872 | + | PROTEIN:ATP-dependent metalloprotease FtsH |
| Dgeo_1833 | Dgeo_1833 | 1946288 | 1947472 | 1185 | - | PROTEIN:MscS mechanosensitive ion channel |
| Dgeo_1834 | Dgeo_1834 | 1947505 | 1948545 | 1041 | - | PROTEIN:putative glycerol-3-phosphate acyltransfer |
| Dgeo_1835 | Dgeo_1835 | 1948529 | 1949113 | 585 | - | PROTEIN:hypothetical protein |
| Dgeo_1836 | Dgeo_1836 | 1949192 | 1949530 | 339 | - | PROTEIN:hypothetical protein |
| Dgeo_1837 | Dgeo_1837 | 1949619 | 1949951 | 333 | + | PROTEIN:thioredoxin |
| Dgeo_1838 | Dgeo_1838 | 1950012 | 1950575 | 564 | - | PROTEIN:hypothetical protein |
| Dgeo_1839 | rplQ | 1950663 | 1951013 | 351 | - | PROTEIN:50S ribosomal protein L17 |
| Dgeo_1840 | Dgeo_1840 | 1951169 | 1952164 | 996 | - | PROTEIN:DNA-directed RNA polymerase subunit alpha |
| Dgeo_1841 | rpsD | 1952178 | 1952798 | 621 | - | PROTEIN:30S ribosomal protein S4 |
| Dgeo_1842 | Dgeo_1842 | 1952941 | 1953336 | 396 | - | PROTEIN:30S ribosomal protein S11 |
| Dgeo_1843 | rpsM | 1953340 | 1953720 | 381 | - | PROTEIN:30S ribosomal protein S13 |
| Dgeo_1844 | rpmJ | 1953723 | 1953836 | 114 | - | PROTEIN:50S ribosomal protein L36 |
| Dgeo_1845 | Dgeo_1845 | 1953955 | 1954254 | 300 | - | PROTEIN:translation initiation factor IF-1 |
| Dgeo_1846 | Dgeo_1846 | 1954347 | 1954940 | 594 | - | PROTEIN:adenylate kinases |
| Dgeo_1847 | secY | 1955095 | 1956420 | 1326 | - | PROTEIN:preprotein translocase subunit SecY |
| Dgeo_1848 | rplO | 1956422 | 1956886 | 465 | - | PROTEIN:50S ribosomal protein L15 |
| Dgeo_1849 | rpmD | 1956883 | 1957071 | 189 | - | PROTEIN:50S ribosomal protein L30 |
| Dgeo_1850 | rpsE | 1957068 | 1957601 | 534 | - | PROTEIN:30S ribosomal protein S5 |
| Dgeo_1851 | rplR | 1957591 | 1957929 | 339 | - | PROTEIN:50S ribosomal protein L18 |
| Dgeo_1852 | rplF | 1957929 | 1958486 | 558 | - | PROTEIN:50S ribosomal protein L6 |
| Dgeo_1853 | rpsH | 1958556 | 1958957 | 402 | - | PROTEIN:30S ribosomal protein S8 |
| Dgeo_1854 | Dgeo_1854 | 1959134 | 1959319 | 186 | - | PROTEIN:ribosomal protein S14 |
| Dgeo_1855 | rplE | 1959345 | 1959884 | 540 | - | PROTEIN:50S ribosomal protein L5 |
| Dgeo_1856 | rplX | 1959946 | 1960293 | 348 | - | PROTEIN:50S ribosomal protein L24 |
| Dgeo_1857 | rplN | 1960293 | 1960697 | 405 | - | PROTEIN:50S ribosomal protein L14 |
| Dgeo_1858 | Dgeo_1858 | 1960694 | 1960978 | 285 | - | PROTEIN:ribosomal protein S17 |
| Dgeo_1859 | Dgeo_1859 | 1960975 | 1961199 | 225 | - | PROTEIN:50S ribosomal protein L29 |
| Dgeo_1860 | rplP | 1961186 | 1961611 | 426 | - | PROTEIN:50S ribosomal protein L16 |
| Dgeo_1861 | rpsC | 1961611 | 1962357 | 747 | - | PROTEIN:30S ribosomal protein S3 |
| Dgeo_1862 | rplV | 1962361 | 1962780 | 420 | - | PROTEIN:50S ribosomal protein L22 |
| Dgeo_1863 | rpsS | 1962777 | 1963064 | 288 | - | PROTEIN:30S ribosomal protein S19 |
| Dgeo_1864 | rplB | 1963077 | 1963910 | 834 | - | PROTEIN:50S ribosomal protein L2 |
| Dgeo_1865 | rplW | 1963924 | 1964211 | 288 | - | PROTEIN:50S ribosomal protein L23 |
| Dgeo_1866 | rplD | 1964208 | 1964849 | 642 | - | PROTEIN:50S ribosomal protein L4 |
| Dgeo_1867 | rplC | 1964853 | 1965476 | 624 | - | PROTEIN:50S ribosomal protein L3 |
| Dgeo_1868 | rpsJ | 1965473 | 1965796 | 324 | - | PROTEIN:30S ribosomal protein S10 |
| Dgeo_1869 | Dgeo_1869 | 1965802 | 1967019 | 1218 | - | PROTEIN:elongation factor Tu |
| Dgeo_1870 | Dgeo_1870 | 1967221 | 1967637 | 417 | - | PROTEIN:hypothetical protein |
| Dgeo_1871 | fusA | 1967745 | 1969835 | 2091 | - | PROTEIN:elongation factor G |
| Dgeo_1872 | Dgeo_1872 | 1969975 | 1970445 | 471 | - | PROTEIN:30S ribosomal protein S7 |
| Dgeo_1873 | rpsL | 1970590 | 1971000 | 411 | - | PROTEIN:30S ribosomal protein S12 |
| Dgeo_1874 | Dgeo_1874 | 1971177 | 1971638 | 462 | - | PROTEIN:hypothetical protein |
| Dgeo_1875 | Dgeo_1875 | 1971635 | 1973278 | 1644 | - | PROTEIN:phosphoglucomutase |
| Dgeo_1876 | glpX | 1973630 | 1974634 | 1005 | + | PROTEIN:fructose 1,6-bisphosphatase II |
| Dgeo_1877 | Dgeo_1877 | 1974777 | 1976123 | 1347 | + | PROTEIN:transposase IS66 |
| Dgeo_1878 | Dgeo_1878 | 1976172 | 1976780 | 609 | + | PROTEIN:flavodoxin/nitric oxide synthase |
| Dgeo_1879 | Dgeo_1879 | 1976796 | 1977545 | 750 | - | PROTEIN:metallophosphoesterase |
| Dgeo_1880 | dcd | 1977597 | 1978157 | 561 | + | PROTEIN:deoxycytidine triphosphate deaminase |
| Dgeo_1881 | Dgeo_1881 | 1978241 | 1978462 | 222 | + | PROTEIN:hypothetical protein |
| Dgeo_1882 | Dgeo_1882 | 1978641 | 1979075 | 435 | - | PROTEIN:thioesterase superfamily protein |
| Dgeo_1883 | Dgeo_1883 | 1979134 | 1980309 | 1176 | + | PROTEIN:Acetyl-CoA C-acetyltransferase |
| Dgeo_1884 | Dgeo_1884 | 1980322 | 1981098 | 777 | + | PROTEIN:ankyrin |
| Dgeo_1885 | Dgeo_1885 | 1981095 | 1981649 | 555 | + | PROTEIN:hypothetical protein |
| Dgeo_1886 | Dgeo_1886 | 1981731 | 1983515 | 1785 | - | PROTEIN:dihydrolipoamide acetyltransferase |
| Dgeo_1887 | aceE | 1983610 | 1986318 | 2709 | - | PROTEIN:pyruvate dehydrogenase subunit E1 |
| Dgeo_1888 | Dgeo_1888 | 1986479 | 1987372 | 894 | + | PROTEIN:LysR family transcriptional regulator |
| Dgeo_1889 | Dgeo_1889 | 1987439 | 1987930 | 492 | + | PROTEIN:DNA topology modulation protein FlaR-relat |
| Dgeo_1890 | Dgeo_1890 | 1988046 | 1990076 | 2031 | + | PROTEIN:excinuclease ABC subunit B |
| Dgeo_1891 | Dgeo_1891 | 1990148 | 1991332 | 1185 | - | PROTEIN:uracil-xanthine permease |
| Dgeo_1892 | Dgeo_1892 | 1991418 | 1991948 | 531 | + | PROTEIN:double-stranded RNA binding |
| Dgeo_1893 | Dgeo_1893 | 1991945 | 1992613 | 669 | + | PROTEIN:HAD family hydrolase |
| Dgeo_1894 | Dgeo_1894 | 1992610 | 1992951 | 342 | + | PROTEIN:hypothetical protein |
| Dgeo_1895 | Dgeo_1895 | 1992948 | 1995407 | 2460 | - | PROTEIN:peptidoglycan glycosyltransferase |
| Dgeo_1896 | Dgeo_1896 | 1995561 | 1996430 | 870 | + | PROTEIN:PP-loop domain-containing protein |
| Dgeo_1897 | Dgeo_1897 | 1996477 | 1996989 | 513 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_1898 | Dgeo_1898 | 1996993 | 1997817 | 825 | - | PROTEIN:hypothetical protein |
| Dgeo_1899 | Dgeo_1899 | 1997879 | 1998298 | 420 | - | PROTEIN:response regulator receiver protein |
| Dgeo_1900 | Dgeo_1900 | 1998630 | 1999421 | 792 | + | PROTEIN:imidazoleglycerol phosphate synthase, cycl |
| Dgeo_1901 | Dgeo_1901 | 1999418 | 2000074 | 657 | + | PROTEIN:bifunctional phosphoribosyl-AMP cyclohydro |
| Dgeo_1902 | Dgeo_1902 | 2000071 | 2000508 | 438 | - | PROTEIN:hypothetical protein |
| Dgeo_1903 | Dgeo_1903 | 2000751 | 2001305 | 555 | + | PROTEIN:5-formyltetrahydrofolate cyclo-ligase |
| Dgeo_1904 | Dgeo_1904 | 2001296 | 2001826 | 531 | - | PROTEIN:hypothetical protein |
| Dgeo_1905 | Dgeo_1905 | 2001903 | 2003027 | 1125 | + | PROTEIN:glycine cleavage system T protein |
| Dgeo_1906 | Dgeo_1906 | 2003124 | 2003486 | 363 | + | PROTEIN:glycine cleavage system H protein |
| Dgeo_1907 | Dgeo_1907 | 2003596 | 2006460 | 2865 | + | PROTEIN:glycine dehydrogenase |
| Dgeo_1908 | Dgeo_1908 | 2006540 | 2007220 | 681 | + | PROTEIN:hypothetical protein |
| Dgeo_1909 | Dgeo_1909 | 2007222 | 2008352 | 1131 | + | PROTEIN:hypothetical protein |
| Dgeo_1910 | Dgeo_1910 | 2008651 | 2010414 | 1764 | + | PROTEIN:DNA primase |
| Dgeo_R0041 | Dgeo_R0041 | 2010487 | 2010562 | 76 | - | RNA:Gly tRNA |
| Dgeo_R0042 | Dgeo_R0042 | 2010573 | 2010689 | 117 | - | RNA:5S ribosomal RNA |
| Dgeo_R0043 | Dgeo_R0043 | 2010773 | 2013662 | 2890 | - | RNA:23S ribosomal RNA |
| Dgeo_1911 | Dgeo_1911 | 2013693 | 2014646 | 954 | + | PROTEIN:periplasmic protein-like protein |
| Dgeo_1912 | Dgeo_1912 | 2014718 | 2016847 | 2130 | - | PROTEIN:hypothetical protein |
| Dgeo_1913 | Dgeo_1913 | 2017004 | 2019091 | 2088 | - | PROTEIN:bifunctional aldehyde dehydrogenase/enoyl- |
| Dgeo_1914 | Dgeo_1914 | 2019124 | 2019867 | 744 | + | PROTEIN:methionine aminopeptidase, type I |
| Dgeo_1915 | Dgeo_1915 | 2019870 | 2020385 | 516 | - | PROTEIN:phenylacetate-CoA oxygenase, PaaJ subunit |
| Dgeo_1916 | Dgeo_1916 | 2020369 | 2021175 | 807 | - | PROTEIN:phenylacetate-CoA oxygenase, PaaI subunit |
| Dgeo_1917 | Dgeo_1917 | 2021172 | 2021750 | 579 | - | PROTEIN:phenylacetic acid degradation B |
| Dgeo_1918 | Dgeo_1918 | 2021750 | 2022712 | 963 | - | PROTEIN:phenylacetate-CoA oxygenase subunit PaaA |
| Dgeo_1919 | Dgeo_1919 | 2022809 | 2023573 | 765 | - | PROTEIN:phosphomethylpyrimidine kinase type-2 |
| Dgeo_1920 | Dgeo_1920 | 2023570 | 2024487 | 918 | - | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_1921 | thiG | 2024549 | 2025352 | 804 | - | PROTEIN:thiazole synthase |
| Dgeo_1922 | Dgeo_1922 | 2025354 | 2025542 | 189 | - | PROTEIN:thiamine biosynthesis protein ThiS |
| Dgeo_1923 | Dgeo_1923 | 2025539 | 2026204 | 666 | - | PROTEIN:thiamine-phosphate pyrophosphorylase |
| Dgeo_1924 | Dgeo_1924 | 2026201 | 2028030 | 1830 | - | PROTEIN:thiamine biosynthesis protein ThiC |
| Dgeo_R0044 | Dgeo_R0044 | 2028181 | 2028298 | 118 | - | RNA:misc_RNA |
| Dgeo_1925 | Dgeo_1925 | 2028384 | 2029373 | 990 | + | PROTEIN:aldo/keto reductase |
| Dgeo_1926 | Dgeo_1926 | 2029384 | 2030778 | 1395 | - | PROTEIN:dienelactone hydrolase-like protein |
| Dgeo_1927 | Dgeo_1927 | 2030913 | 2031317 | 405 | + | PROTEIN:phosphopantethiene-protein transferase dom |
| Dgeo_1928 | Dgeo_1928 | 2031373 | 2032992 | 1620 | - | PROTEIN:major facilitator transporter |
| Dgeo_1929 | Dgeo_1929 | 2032989 | 2033468 | 480 | - | PROTEIN:MarR family transcriptional regulator |
| Dgeo_1930 | Dgeo_1930 | 2033519 | 2033974 | 456 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_1932 | Dgeo_1932 | 2035035 | 2036135 | 1101 | - | PROTEIN:major facilitator transporter |
| Dgeo_R0045 | Dgeo_R0045 | 2036288 | 2036401 | 114 | + | RNA:misc_RNA |
| Dgeo_1933 | Dgeo_1933 | 2036459 | 2037490 | 1032 | + | PROTEIN:ABC transporter periplasmic-binding protei |
| Dgeo_1934 | Dgeo_1934 | 2037487 | 2039073 | 1587 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_1935 | Dgeo_1935 | 2039066 | 2039650 | 585 | - | PROTEIN:hypothetical protein |
| Dgeo_1936 | Dgeo_1936 | 2039709 | 2039975 | 267 | + | PROTEIN:transcriptional regulator/antitoxin, MazE |
| Dgeo_1937 | Dgeo_1937 | 2039972 | 2040304 | 333 | + | PROTEIN:transcriptional modulator of MazE/toxin, M |
| Dgeo_1938 | Dgeo_1938 | 2040301 | 2041278 | 978 | + | PROTEIN:ABC transporter related |
| Dgeo_1939 | Dgeo_1939 | 2041289 | 2041927 | 639 | + | PROTEIN:thiamine pyrophosphokinase |
| Dgeo_1940 | Dgeo_1940 | 2041937 | 2043340 | 1404 | - | PROTEIN:hypothetical protein |
| Dgeo_1941 | Dgeo_1941 | 2043525 | 2044184 | 660 | + | PROTEIN:hypothetical protein |
| Dgeo_1942 | Dgeo_1942 | 2044257 | 2045630 | 1374 | + | PROTEIN:glutamate-1-semialdehyde-2,1-aminomutase |
| Dgeo_1943 | Dgeo_1943 | 2045627 | 2046055 | 429 | + | PROTEIN:CoA-binding |
| Dgeo_1944 | Dgeo_1944 | 2046132 | 2047328 | 1197 | - | PROTEIN:MoeA-like protein |
| Dgeo_1945 | Dgeo_1945 | 2047469 | 2047948 | 480 | + | PROTEIN:peptidylprolyl isomerase, FKBP-type |
| Dgeo_1946 | Dgeo_1946 | 2048035 | 2048826 | 792 | - | PROTEIN:molybdopterin dehydrogenase, FAD-binding |
| Dgeo_1947 | Dgeo_1947 | 2048844 | 2051234 | 2391 | - | PROTEIN:aldehyde oxidase and xanthine dehydrogenas |
| Dgeo_1948 | Dgeo_1948 | 2051258 | 2051755 | 498 | - | PROTEIN:(2Fe-2S)-binding |
| Dgeo_1949 | Dgeo_1949 | 2052083 | 2053222 | 1140 | + | PROTEIN:NADH dehydrogenase |
| Dgeo_1950 | Dgeo_1950 | 2053296 | 2053868 | 573 | + | PROTEIN:hypothetical protein |
| Dgeo_1951 | valS | 2054182 | 2056959 | 2778 | + | PROTEIN:valyl-tRNA synthetase |
| Dgeo_1952 | Dgeo_1952 | 2057028 | 2058182 | 1155 | - | PROTEIN:aminotransferase, class V |
| Dgeo_1953 | Dgeo_1953 | 2058213 | 2059895 | 1683 | - | PROTEIN:PASTA domain-containing protein |
| Dgeo_1954 | Dgeo_1954 | 2060011 | 2060307 | 297 | - | PROTEIN:branched-chain amino acid transport |
| Dgeo_1955 | Dgeo_1955 | 2060304 | 2061032 | 729 | - | PROTEIN:AzlC-like protein |
| Dgeo_1956 | Dgeo_1956 | 2061345 | 2061680 | 336 | + | PROTEIN:small multidrug resistance protein |
| Dgeo_1957 | Dgeo_1957 | 2061677 | 2062015 | 339 | + | PROTEIN:small multidrug resistance protein |
| Dgeo_1958 | Dgeo_1958 | 2062029 | 2062724 | 696 | - | PROTEIN:amino acid ABC transporter permease |
| Dgeo_1959 | Dgeo_1959 | 2062787 | 2063455 | 669 | - | PROTEIN:extracellular solute-binding protein |
| Dgeo_1960 | Dgeo_1960 | 2063549 | 2064040 | 492 | + | PROTEIN:hypothetical protein |
| Dgeo_1961 | Dgeo_1961 | 2064265 | 2064648 | 384 | - | PROTEIN:hypothetical protein |
| Dgeo_1962 | Dgeo_1962 | 2064653 | 2065153 | 501 | - | PROTEIN:hypothetical protein |
| Dgeo_1963 | Dgeo_1963 | 2065255 | 2065992 | 738 | + | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_1964 | Dgeo_1964 | 2066180 | 2066458 | 279 | + | PROTEIN:twin arginine-targeting protein translocas |
| Dgeo_1965 | Dgeo_1965 | 2066471 | 2067241 | 771 | + | PROTEIN:Sec-independent protein translocase TatC |
| Dgeo_1966 | Dgeo_1966 | 2067336 | 2068247 | 912 | + | PROTEIN:prolipoprotein diacylglyceryl transferase |
| Dgeo_1967 | Dgeo_1967 | 2068365 | 2069810 | 1446 | + | PROTEIN:UDP-N-acetylglucosamine pyrophosphorylase |
| Dgeo_1968 | Dgeo_1968 | 2069916 | 2071301 | 1386 | - | PROTEIN:major facilitator transporter |
| Dgeo_1969 | Dgeo_1969 | 2071676 | 2072131 | 456 | - | PROTEIN:ribonuclease H |
| Dgeo_1970 | Dgeo_1970 | 2072277 | 2072765 | 489 | + | PROTEIN:hypothetical protein |
| Dgeo_1971 | Dgeo_1971 | 2072870 | 2073619 | 750 | + | PROTEIN:LmbE-like protein protein |
| Dgeo_1972 | Dgeo_1972 | 2073687 | 2074238 | 552 | + | PROTEIN:hypothetical protein |
| Dgeo_1973 | Dgeo_1973 | 2074413 | 2075495 | 1083 | + | PROTEIN:6-phosphogluconate dehydrogenase-like prot |
| Dgeo_1974 | Dgeo_1974 | 2075492 | 2077174 | 1683 | + | PROTEIN:glucose-6-phosphate 1-dehydrogenase |
| Dgeo_1975 | Dgeo_1975 | 2077185 | 2078156 | 972 | + | PROTEIN:glucose 6-phosphate dehydrogenase assembly |
| Dgeo_1976 | Dgeo_1976 | 2078153 | 2078830 | 678 | + | PROTEIN:6-phosphogluconolactonase |
| Dgeo_1977 | Dgeo_1977 | 2078981 | 2081767 | 2787 | + | PROTEIN:peptidase M16-like protein |
| Dgeo_1978 | Dgeo_1978 | 2081814 | 2083007 | 1194 | + | PROTEIN:hypothetical protein |
| Dgeo_1979 | Dgeo_1979 | 2083049 | 2083429 | 381 | + | PROTEIN:hypothetical protein |
| Dgeo_1980 | Dgeo_1980 | 2083469 | 2084338 | 870 | + | PROTEIN:glutamine cyclotransferase |
| Dgeo_1981 | Dgeo_1981 | 2084394 | 2084792 | 399 | + | PROTEIN:hypothetical protein |
| Dgeo_1982 | Dgeo_1982 | 2084807 | 2085607 | 801 | + | PROTEIN:hypothetical protein |
| Dgeo_1983 | Dgeo_1983 | 2085684 | 2086316 | 633 | + | PROTEIN:thymidylate kinase |
| Dgeo_1984 | Dgeo_1984 | 2086395 | 2087648 | 1254 | + | PROTEIN:hypothetical protein |
| Dgeo_R0046 | Dgeo_R0046 | 2087728 | 2087804 | 77 | - | RNA:Met tRNA |
| Dgeo_R0047 | Dgeo_R0047 | 2087843 | 2087918 | 76 | - | RNA:Ala tRNA |
| Dgeo_1985 | Dgeo_1985 | 2087956 | 2088630 | 675 | - | PROTEIN:transcriptional regulator, TrmB |
| Dgeo_1986 | Dgeo_1986 | 2088800 | 2089561 | 762 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_1987 | Dgeo_1987 | 2089672 | 2090343 | 672 | + | PROTEIN:amino acid ABC transporter permease |
| Dgeo_R0048 | Dgeo_R0048 | 2090397 | 2090471 | 75 | - | RNA:Val tRNA |
| Dgeo_R0049 | Dgeo_R0049 | 2090498 | 2090573 | 76 | - | RNA:Met tRNA |
| Dgeo_1988 | Dgeo_1988 | 2090639 | 2091982 | 1344 | + | PROTEIN:glycogen/starch synthases, ADP-glucose typ |
| Dgeo_1989 | Dgeo_1989 | 2091986 | 2092693 | 708 | - | PROTEIN:alpha/beta hydrolase fold |
| Dgeo_1990 | Dgeo_1990 | 2092827 | 2093591 | 765 | + | PROTEIN:RNA methyltransferase TrmH, group 1 |
| Dgeo_1991 | Dgeo_1991 | 2093598 | 2093837 | 240 | - | PROTEIN:hypothetical protein |
| Dgeo_1992 | Dgeo_1992 | 2093902 | 2095626 | 1725 | + | PROTEIN:multi-sensor signal transduction histidine |
| Dgeo_1993 | apaG | 2095686 | 2096078 | 393 | - | PROTEIN:ApaG |
| Dgeo_1994 | Dgeo_1994 | 2096115 | 2097179 | 1065 | + | PROTEIN:peptidase M42 |
| Dgeo_1995 | Dgeo_1995 | 2097321 | 2097752 | 432 | + | PROTEIN:hypothetical protein |
| Dgeo_1996 | Dgeo_1996 | 2097811 | 2098845 | 1035 | - | PROTEIN:tRNA-dihydrouridine synthase A |
| Dgeo_1997 | Dgeo_1997 | 2098965 | 2099270 | 306 | + | PROTEIN:hypothetical protein |
| Dgeo_1998 | Dgeo_1998 | 2099344 | 2100525 | 1182 | - | PROTEIN:hypothetical protein |
| Dgeo_1999 | Dgeo_1999 | 2100601 | 2104314 | 3714 | - | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_2000 | Dgeo_2000 | 2104370 | 2104696 | 327 | - | PROTEIN:hypothetical protein |
| Dgeo_2001 | Dgeo_2001 | 2104701 | 2107595 | 2895 | - | PROTEIN:DNA topoisomerase I |
| Dgeo_2002 | Dgeo_2002 | 2107714 | 2108310 | 597 | + | PROTEIN:hypothetical protein |
| Dgeo_2003 | Dgeo_2003 | 2108321 | 2109937 | 1617 | - | PROTEIN:integral membrane protein MviN |
| Dgeo_2004 | Dgeo_2004 | 2109974 | 2110114 | 141 | - | PROTEIN:hypothetical protein |
| Dgeo_2005 | Dgeo_2005 | 2110111 | 2110980 | 870 | - | PROTEIN:hypothetical protein |
| Dgeo_R0050 | Dgeo_R0050 | 2111082 | 2111171 | 90 | + | RNA:Ser tRNA |
| Dgeo_2006 | Dgeo_2006 | 2111471 | 2113180 | 1710 | + | PROTEIN:GTP-binding protein, HSR1-related |
| Dgeo_2007 | Dgeo_2007 | 2113260 | 2114267 | 1008 | - | PROTEIN:transposase, IS4 |
| Dgeo_2008 | Dgeo_2008 | 2114828 | 2116999 | 2172 | + | PROTEIN:dTDP-4-dehydrorhamnose reductase |
| Dgeo_2009 | Dgeo_2009 | 2117060 | 2118013 | 954 | + | PROTEIN:diacylglycerol kinase, catalytic region |
| Dgeo_2010 | Dgeo_2010 | 2118016 | 2118759 | 744 | + | PROTEIN:PHP-like protein |
| Dgeo_R0051 | Dgeo_R0051 | 2118878 | 2119225 | 348 | + | RNA:tmRNA |
| Dgeo_2011 | Dgeo_2011 | 2119343 | 2119822 | 480 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2012 | Dgeo_2012 | 2119917 | 2123057 | 3141 | - | PROTEIN:HsdR family type I site-specific deoxyribo |
| Dgeo_2013 | Dgeo_2013 | 2123062 | 2124054 | 993 | - | PROTEIN:death-on-curing protein |
| Dgeo_2014 | Dgeo_2014 | 2124051 | 2124377 | 327 | - | PROTEIN:type I restriction-modification system end |
| Dgeo_2015 | Dgeo_2015 | 2124410 | 2126179 | 1770 | - | PROTEIN:hypothetical protein |
| Dgeo_2016 | Dgeo_2016 | 2126176 | 2127429 | 1254 | - | PROTEIN:restriction modification system DNA specif |
| Dgeo_2017 | Dgeo_2017 | 2127419 | 2128972 | 1554 | - | PROTEIN:N-6 DNA methylase |
| Dgeo_R0052 | Dgeo_R0052 | 2129216 | 2129302 | 87 | + | RNA:Leu tRNA |
| Dgeo_2018 | Dgeo_2018 | 2129334 | 2129759 | 426 | - | PROTEIN:hypothetical protein |
| Dgeo_2019 | Dgeo_2019 | 2129783 | 2130835 | 1053 | - | PROTEIN:AFG1-like protein ATPase |
| Dgeo_2020 | Dgeo_2020 | 2130860 | 2131648 | 789 | - | PROTEIN:RNA-binding S4 |
| Dgeo_2021 | Dgeo_2021 | 2131707 | 2132093 | 387 | + | PROTEIN:hypothetical protein |
| Dgeo_2022 | Dgeo_2022 | 2132114 | 2132854 | 741 | + | PROTEIN:RNA methyltransferase TrmH, group 3 |
| Dgeo_2023 | Dgeo_2023 | 2132851 | 2133747 | 897 | + | PROTEIN:aldose 1-epimerase |
| Dgeo_2024 | Dgeo_2024 | 2133852 | 2134070 | 219 | - | PROTEIN:hypothetical protein |
| Dgeo_2025 | Dgeo_2025 | 2134119 | 2135246 | 1128 | - | PROTEIN:peptidase S1 and S6, chymotrypsin/Hap |
| Dgeo_2026 | Dgeo_2026 | 2135252 | 2135821 | 570 | + | PROTEIN:hypothetical protein |
| Dgeo_2027 | Dgeo_2027 | 2135833 | 2136534 | 702 | + | PROTEIN:thymidylate synthase (FAD) |
| Dgeo_2028 | Dgeo_2028 | 2136672 | 2137430 | 759 | + | PROTEIN:ABC transporter related |
| Dgeo_2029 | Dgeo_2029 | 2137427 | 2138167 | 741 | + | PROTEIN:ABC transporter related |
| Dgeo_2030 | Dgeo_2030 | 2138189 | 2139412 | 1224 | + | PROTEIN:LivK, ABC-type branched-chain amino acid t |
| Dgeo_2031 | Dgeo_2031 | 2139481 | 2140383 | 903 | + | PROTEIN:inner-membrane translocator |
| Dgeo_2032 | Dgeo_2032 | 2140380 | 2141336 | 957 | + | PROTEIN:inner-membrane translocator |
| Dgeo_2033 | Dgeo_2033 | 2141382 | 2141834 | 453 | - | PROTEIN:transcriptional regulator NrdR |
| Dgeo_2034 | Dgeo_2034 | 2141965 | 2142678 | 714 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_2035 | Dgeo_2035 | 2142725 | 2142994 | 270 | - | PROTEIN:hypothetical protein |
| Dgeo_2036 | Dgeo_2036 | 2143015 | 2143701 | 687 | - | PROTEIN:NUDIX hydrolase |
| Dgeo_2037 | Dgeo_2037 | 2143701 | 2146250 | 2550 | - | PROTEIN:DnaB-like protein helicase |
| Dgeo_2038 | Dgeo_2038 | 2146607 | 2146996 | 390 | + | PROTEIN:fimbrial protein pilin |
| Dgeo_2039 | Dgeo_2039 | 2147059 | 2148447 | 1389 | + | PROTEIN:O-antigen polymerase |
| Dgeo_2040 | Dgeo_2040 | 2148573 | 2149139 | 567 | + | PROTEIN:hypothetical protein |
| Dgeo_2041 | Dgeo_2041 | 2149202 | 2149672 | 471 | - | PROTEIN:hypothetical protein |
| Dgeo_2042 | Dgeo_2042 | 2149851 | 2150621 | 771 | + | PROTEIN:methyltransferase type 11 |
| Dgeo_2043 | Dgeo_2043 | 2150618 | 2151598 | 981 | + | PROTEIN:glycerol-3-phosphate dehydrogenase (NAD(P) |
| Dgeo_2044 | Dgeo_2044 | 2151658 | 2151888 | 231 | + | PROTEIN:hypothetical protein |
| Dgeo_2045 | Dgeo_2045 | 2152033 | 2152701 | 669 | - | PROTEIN:V-type ATPase, D subunit |
| Dgeo_2046 | Dgeo_2046 | 2152774 | 2154186 | 1413 | - | PROTEIN:V-type ATP synthase subunit B |
| Dgeo_2047 | Dgeo_2047 | 2154183 | 2155931 | 1749 | - | PROTEIN:V-type ATP synthase subunit A |
| Dgeo_2048 | Dgeo_2048 | 2156014 | 2156352 | 339 | - | PROTEIN:vacuolar H+-transporting two-sector ATPase |
| Dgeo_2049 | Dgeo_2049 | 2156349 | 2157326 | 978 | - | PROTEIN:H+-transporting two-sector ATPase, C (AC39 |
| Dgeo_2050 | Dgeo_2050 | 2157338 | 2157895 | 558 | - | PROTEIN:H+-transporting two-sector ATPase, E subun |
| Dgeo_2051 | Dgeo_2051 | 2157897 | 2158205 | 309 | - | PROTEIN:H+-transporting two-sector ATPase, C subun |
| Dgeo_2052 | Dgeo_2052 | 2158225 | 2160294 | 2070 | - | PROTEIN:V-type ATPase, 116 kDa subunit |
| Dgeo_2053 | Dgeo_2053 | 2160291 | 2160632 | 342 | - | PROTEIN:hypothetical protein |
| Dgeo_2054 | Dgeo_2054 | 2160859 | 2162154 | 1296 | - | PROTEIN:ammonium transporter |
| Dgeo_2055 | Dgeo_2055 | 2162151 | 2162543 | 393 | - | PROTEIN:nitrogen regulatory protein P-II |
| Dgeo_2056 | Dgeo_2056 | 2162884 | 2163528 | 645 | - | PROTEIN:peptidyl-tRNA hydrolase |
| Dgeo_2057 | Dgeo_2057 | 2164079 | 2165191 | 1113 | + | PROTEIN:(S)-2-hydroxy-acid oxidase |
| Dgeo_2058 | Dgeo_2058 | 2165243 | 2166292 | 1050 | - | PROTEIN:type I topoisomerase, putative |
| Dgeo_2059 | Dgeo_2059 | 2166292 | 2167029 | 738 | - | PROTEIN:uracil-DNA glycosylase |
| Dgeo_2060 | Dgeo_2060 | 2167114 | 2168298 | 1185 | - | PROTEIN:carbamoyl phosphate synthase small subunit |
| Dgeo_2061 | Dgeo_2061 | 2168442 | 2168972 | 531 | - | PROTEIN:acetyltransferase |
| Dgeo_2062 | Dgeo_2062 | 2169037 | 2169507 | 471 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2063 | Dgeo_2063 | 2169624 | 2170130 | 507 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2064 | Dgeo_2064 | 2170158 | 2170652 | 495 | - | PROTEIN:histidine triad (HIT) protein |
| Dgeo_2065 | Dgeo_2065 | 2170661 | 2172058 | 1398 | - | PROTEIN:argininosuccinate lyase |
| Dgeo_2066 | Dgeo_2066 | 2172120 | 2172590 | 471 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2067 | Dgeo_2067 | 2172594 | 2173838 | 1245 | - | PROTEIN:argininosuccinate synthase |
| Dgeo_2068 | Dgeo_2068 | 2174114 | 2174578 | 465 | - | PROTEIN:hypothetical protein |
| Dgeo_2069 | Dgeo_2069 | 2174647 | 2175774 | 1128 | - | PROTEIN:aminotransferase, class I and II |
| Dgeo_2070 | carB | 2176096 | 2179185 | 3090 | + | PROTEIN:carbamoyl phosphate synthase large subunit |
| Dgeo_2071 | Dgeo_2071 | 2179418 | 2180713 | 1296 | + | PROTEIN:sodium:dicarboxylate symporter |
| Dgeo_2072 | Dgeo_2072 | 2180959 | 2181417 | 459 | - | PROTEIN:methionine sulfoxide reductase B |
| Dgeo_2073 | Dgeo_2073 | 2181526 | 2182296 | 771 | + | PROTEIN:DSBA oxidoreductase |
| Dgeo_2074 | Dgeo_2074 | 2182315 | 2183928 | 1614 | - | PROTEIN:carboxylesterase, type B |
| Dgeo_2075 | Dgeo_2075 | 2183992 | 2185857 | 1866 | - | PROTEIN:ATP-dependent metalloprotease FtsH |
| Dgeo_2076 | Dgeo_2076 | 2186109 | 2187998 | 1890 | + | PROTEIN:chaperone DnaK |
| Dgeo_2077 | Dgeo_2077 | 2188111 | 2188767 | 657 | + | PROTEIN:GrpE protein |
| Dgeo_2078 | Dgeo_2078 | 2188854 | 2189762 | 909 | + | PROTEIN:chaperone DnaJ-like protein |
| Dgeo_2079 | Dgeo_2079 | 2189789 | 2190031 | 243 | + | PROTEIN:hypothetical protein |
| Dgeo_2080 | Dgeo_2080 | 2190079 | 2190504 | 426 | - | PROTEIN:signal-transduction protein |
| Dgeo_2081 | Dgeo_2081 | 2190618 | 2193164 | 2547 | - | PROTEIN:peptidase S8 and S53, subtilisin, kexin, s |
| Dgeo_2082 | Dgeo_2082 | 2193292 | 2194104 | 813 | - | PROTEIN:hypothetical protein |
| Dgeo_2083 | Dgeo_2083 | 2194170 | 2195096 | 927 | - | PROTEIN:fructose-1,6-bisphosphate aldolase, class |
| Dgeo_2084 | Dgeo_2084 | 2195200 | 2196420 | 1221 | + | PROTEIN:aminotransferase, class I and II |
| Dgeo_2085 | Dgeo_2085 | 2196443 | 2197258 | 816 | + | PROTEIN:glutamate racemase |
| Dgeo_2086 | Dgeo_2086 | 2197255 | 2197968 | 714 | + | PROTEIN:ribonuclease PH |
| Dgeo_2087 | Dgeo_2087 | 2198053 | 2198412 | 360 | + | PROTEIN:DoxX |
| Dgeo_2088 | Dgeo_2088 | 2198521 | 2198727 | 207 | + | PROTEIN:hypothetical protein |
| Dgeo_2089 | Dgeo_2089 | 2198744 | 2199691 | 948 | - | PROTEIN:methyltransferase type 12 |
| Dgeo_2090 | Dgeo_2090 | 2199696 | 2200691 | 996 | - | PROTEIN:tRNA (guanine-N(7)-)-methyltransferase |
| Dgeo_2091 | Dgeo_2091 | 2200688 | 2201431 | 744 | - | PROTEIN:glucose-inhibited division protein A |
| Dgeo_2092 | Dgeo_2092 | 2201524 | 2202519 | 996 | - | PROTEIN:metallo-beta-lactamase superfamily hydrola |
| Dgeo_2093 | Dgeo_2093 | 2202516 | 2202884 | 369 | - | PROTEIN:hypothetical protein |
| Dgeo_2094 | Dgeo_2094 | 2202975 | 2203433 | 459 | + | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2095 | Dgeo_2095 | 2203396 | 2203821 | 426 | - | PROTEIN:response regulator receiver protein |
| Dgeo_2096 | Dgeo_2096 | 2203818 | 2207240 | 3423 | - | PROTEIN:multi-sensor signal transduction histidine |
| Dgeo_2097 | Dgeo_2097 | 2207310 | 2208440 | 1131 | + | PROTEIN:hypothetical protein |
| Dgeo_2098 | Dgeo_2098 | 2208442 | 2208795 | 354 | + | PROTEIN:hypothetical protein |
| Dgeo_2099 | Dgeo_2099 | 2208827 | 2209711 | 885 | + | PROTEIN:hypothetical protein |
| Dgeo_2100 | Dgeo_2100 | 2209708 | 2210193 | 486 | + | PROTEIN:DinB |
| Dgeo_2101 | Dgeo_2101 | 2210186 | 2210560 | 375 | + | PROTEIN:methylated-DNA-(protein)-cysteine S-methyl |
| Dgeo_2102 | Dgeo_2102 | 2210634 | 2211440 | 807 | + | PROTEIN:cytochrome c, class I |
| Dgeo_2103 | engA | 2211813 | 2213138 | 1326 | + | PROTEIN:GTP-binding protein EngA |
| Dgeo_2104 | Dgeo_2104 | 2213592 | 2215475 | 1884 | - | PROTEIN:glycoside hydrolase family protein |
| Dgeo_2105 | Dgeo_2105 | 2215735 | 2216013 | 279 | - | PROTEIN:hypothetical protein |
| Dgeo_R0053 | Dgeo_R0053 | 2216152 | 2216226 | 75 | - | RNA:Thr tRNA |
| Dgeo_2106 | Dgeo_2106 | 2216320 | 2218014 | 1695 | - | PROTEIN:dynamin |
| Dgeo_2107 | Dgeo_2107 | 2218073 | 2218405 | 333 | - | PROTEIN:hypothetical protein |
| Dgeo_2108 | Dgeo_2108 | 2218386 | 2219477 | 1092 | - | PROTEIN:transposase, IS4 |
| Dgeo_2109 | Dgeo_2109 | 2219515 | 2220015 | 501 | - | PROTEIN:hypothetical protein |
| Dgeo_2110 | Dgeo_2110 | 2220012 | 2220521 | 510 | - | PROTEIN:putative pilin, type IV |
| Dgeo_2111 | Dgeo_2111 | 2220518 | 2220997 | 480 | - | PROTEIN:putative pilin, type IV |
| Dgeo_2112 | Dgeo_2112 | 2220994 | 2223075 | 2082 | - | PROTEIN:hypothetical protein |
| Dgeo_2113 | Dgeo_2113 | 2223216 | 2223428 | 213 | + | PROTEIN:hypothetical protein |
| Dgeo_2114 | Dgeo_2114 | 2223444 | 2223758 | 315 | - | PROTEIN:hypothetical protein |
| Dgeo_2115 | Dgeo_2115 | 2223963 | 2224820 | 858 | - | PROTEIN:protein of unknown function, zinc metallop |
| Dgeo_2116 | Dgeo_2116 | 2224994 | 2228410 | 3417 | - | PROTEIN:acriflavin resistance protein |
| Dgeo_2117 | Dgeo_2117 | 2228407 | 2229693 | 1287 | - | PROTEIN:secretion protein HlyD |
| Dgeo_2118 | Dgeo_2118 | 2229699 | 2230736 | 1038 | - | PROTEIN:hypothetical protein |
| Dgeo_2119 | Dgeo_2119 | 2230733 | 2232169 | 1437 | - | PROTEIN:outer membrane efflux protein |
| Dgeo_2120 | Dgeo_2120 | 2232243 | 2232830 | 588 | - | PROTEIN:TetR family transcriptional regulator |
| Dgeo_2121 | Dgeo_2121 | 2232948 | 2233967 | 1020 | - | PROTEIN:D-alanine--D-alanine ligase |
| Dgeo_2122 | Dgeo_2122 | 2234162 | 2235883 | 1722 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_2123 | Dgeo_2123 | 2236046 | 2237068 | 1023 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_2124 | Dgeo_2124 | 2237068 | 2238090 | 1023 | + | PROTEIN:binding-protein-dependent transport system |
| Dgeo_2125 | Dgeo_2125 | 2238087 | 2238662 | 576 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2126 | Dgeo_2126 | 2238673 | 2239164 | 492 | - | PROTEIN:hypothetical protein |
| Dgeo_2127 | Dgeo_2127 | 2239157 | 2240053 | 897 | - | PROTEIN:glycine cleavage T protein (aminomethyl tr |
| Dgeo_2128 | Dgeo_2128 | 2240116 | 2241213 | 1098 | - | PROTEIN:glutamyl-tRNA reductase |
| Dgeo_2129 | Dgeo_2129 | 2241210 | 2241776 | 567 | - | PROTEIN:siroheme synthase |
| Dgeo_2130 | Dgeo_2130 | 2241748 | 2242047 | 300 | - | PROTEIN:hypothetical protein |
| Dgeo_2131 | Dgeo_2131 | 2242346 | 2243869 | 1524 | - | PROTEIN:uroporphyrin-III C-methyltransferase |
| Dgeo_2132 | Dgeo_2132 | 2243953 | 2244639 | 687 | - | PROTEIN:hypothetical protein |
| Dgeo_2133 | Dgeo_2133 | 2244765 | 2245670 | 906 | - | PROTEIN:peptidase S58, DmpA |
| Dgeo_2134 | Dgeo_2134 | 2245804 | 2247414 | 1611 | - | PROTEIN:hypothetical protein |
| Dgeo_2135 | Dgeo_2135 | 2247488 | 2249701 | 2214 | - | PROTEIN:DNA polymerase III, subunits gamma and tau |
| Dgeo_2136 | Dgeo_2136 | 2250115 | 2251335 | 1221 | + | PROTEIN:competence-damaged protein |
| Dgeo_2137 | Dgeo_2137 | 2251332 | 2251994 | 663 | + | PROTEIN:2',5' RNA ligase |
| Dgeo_2138 | recA | 2251991 | 2253067 | 1077 | + | PROTEIN:recombinase A |
| Dgeo_2139 | Dgeo_2139 | 2253261 | 2253839 | 579 | - | PROTEIN:BioY protein |
| Dgeo_2140 | Dgeo_2140 | 2253836 | 2254810 | 975 | - | PROTEIN:bifunctional BirA, biotin operon repressor |
| Dgeo_2141 | Dgeo_2141 | 2254920 | 2255333 | 414 | + | PROTEIN:ferric uptake regulator family protein |
| Dgeo_2142 | Dgeo_2142 | 2255387 | 2256040 | 654 | + | PROTEIN:hypothetical protein |
| Dgeo_2143 | Dgeo_2143 | 2256076 | 2256720 | 645 | + | PROTEIN:multiple antibiotic resistance (MarC)-rela |
| Dgeo_2144 | Dgeo_2144 | 2256797 | 2257366 | 570 | + | PROTEIN:HNH endonuclease |
| Dgeo_2145 | Dgeo_2145 | 2257439 | 2258002 | 564 | + | PROTEIN:metal dependent phosphohydrolase |
| Dgeo_2146 | Dgeo_2146 | 2258145 | 2258681 | 537 | + | PROTEIN:hypothetical protein |
| Dgeo_2147 | Dgeo_2147 | 2258753 | 2259214 | 462 | - | PROTEIN:response regulator receiver protein |
| Dgeo_2148 | Dgeo_2148 | 2259220 | 2261532 | 2313 | - | PROTEIN:PAS/PAC sensor signal transduction histidi |
| Dgeo_2149 | Dgeo_2149 | 2261548 | 2262213 | 666 | - | PROTEIN:two component transcriptional regulator |
| Dgeo_2150 | Dgeo_2150 | 2262491 | 2264182 | 1692 | + | PROTEIN:beta-lactamase-like protein |
| Dgeo_2151 | Dgeo_2151 | 2264334 | 2264936 | 603 | + | PROTEIN:endopeptidase Clp |
| Dgeo_2152 | clpX | 2264933 | 2266153 | 1221 | + | PROTEIN:ATP-dependent protease ATP-binding subunit |
| Dgeo_2153 | Dgeo_2153 | 2266327 | 2268768 | 2442 | + | PROTEIN:ATP-dependent protease La |
| Dgeo_2154 | Dgeo_2154 | 2268909 | 2270645 | 1737 | - | PROTEIN:malate dehydrogenase |
| Dgeo_2155 | Dgeo_2155 | 2270701 | 2272449 | 1749 | - | PROTEIN:malate dehydrogenase |
| Dgeo_2156 | Dgeo_2156 | 2272526 | 2272945 | 420 | - | PROTEIN:thioesterase-like protein |
| Dgeo_2157 | Dgeo_2157 | 2273067 | 2274248 | 1182 | + | PROTEIN:ribonuclease BN |
| Dgeo_2158 | Dgeo_2158 | 2274277 | 2275008 | 732 | - | PROTEIN:phosphoribosylformimino-5-aminoimidazole c |
| Dgeo_2159 | Dgeo_2159 | 2275074 | 2275871 | 798 | - | PROTEIN:MerR family transcriptional regulator |
| Dgeo_2160 | Dgeo_2160 | 2275878 | 2277263 | 1386 | + | PROTEIN:glycosyl transferase, group 1 |
| Dgeo_R0054 | Dgeo_R0054 | 2277302 | 2277386 | 85 | - | RNA:Leu tRNA |
| Dgeo_2161 | Dgeo_2161 | 2277542 | 2278546 | 1005 | - | PROTEIN:malate dehydrogenase |
| Dgeo_2162 | Dgeo_2162 | 2278701 | 2279522 | 822 | - | PROTEIN:enoyl-CoA hydratase |
| Dgeo_2163 | Dgeo_2163 | 2279934 | 2280530 | 597 | - | PROTEIN:hypothetical protein |
| Dgeo_2164 | Dgeo_2164 | 2280616 | 2281146 | 531 | - | PROTEIN:thioesterase superfamily protein |
| Dgeo_2165 | Dgeo_2165 | 2281263 | 2282396 | 1134 | + | PROTEIN:extracellular ligand-binding receptor |
| Dgeo_2166 | Dgeo_2166 | 2282472 | 2283332 | 861 | + | PROTEIN:inner-membrane translocator |
| Dgeo_2167 | Dgeo_2167 | 2283329 | 2284306 | 978 | + | PROTEIN:inner-membrane translocator |
| Dgeo_2168 | Dgeo_2168 | 2284299 | 2285123 | 825 | + | PROTEIN:ABC transporter related |
| Dgeo_2169 | Dgeo_2169 | 2285116 | 2285823 | 708 | + | PROTEIN:ABC transporter related |
| Dgeo_2170 | Dgeo_2170 | 2285966 | 2286286 | 321 | + | PROTEIN:small multidrug resistance protein |
| Dgeo_2171 | Dgeo_2171 | 2286336 | 2287310 | 975 | - | PROTEIN:aldo/keto reductase |
| Dgeo_2172 | Dgeo_2172 | 2287455 | 2288267 | 813 | - | PROTEIN:DeoR family transcriptional regulator |
| Dgeo_2173 | Dgeo_2173 | 2288502 | 2291009 | 2508 | + | PROTEIN:phosphoenolpyruvate-protein phosphotransfe |
| Dgeo_2174 | Dgeo_2174 | 2291006 | 2291986 | 981 | + | PROTEIN:1-phosphofructokinase |
| Dgeo_2175 | Dgeo_2175 | 2292051 | 2293874 | 1824 | + | PROTEIN:phosphotransferase system, fructose IIC co |
| Dgeo_2176 | Dgeo_2176 | 2294035 | 2294955 | 921 | - | PROTEIN:acetyl-CoA carboxylase, carboxyl transfera |
| Dgeo_2177 | Dgeo_2177 | 2294952 | 2295806 | 855 | - | PROTEIN:acetyl-CoA carboxylase, carboxyl transfera |
| Dgeo_2178 | Dgeo_2178 | 2295927 | 2296379 | 453 | - | PROTEIN:hypothetical protein |
| Dgeo_2179 | Dgeo_2179 | 2296437 | 2298569 | 2133 | - | PROTEIN:multi-sensor signal transduction histidine |
| Dgeo_2180 | Dgeo_2180 | 2298605 | 2299912 | 1308 | - | PROTEIN:pyrimidine-nucleoside phosphorylase |
| Dgeo_2181 | Dgeo_2181 | 2300213 | 2300470 | 258 | + | PROTEIN:hypothetical protein |
| Dgeo_2182 | Dgeo_2182 | 2300574 | 2302439 | 1866 | - | PROTEIN:ATP-dependent metalloprotease FtsH |
| Dgeo_2183 | Dgeo_2183 | 2302648 | 2304048 | 1401 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_2184 | Dgeo_2184 | 2304045 | 2304494 | 450 | - | PROTEIN:hypothetical protein |
| Dgeo_2185 | Dgeo_2185 | 2304624 | 2305778 | 1155 | + | PROTEIN:peptidase S1 and S6, chymotrypsin/Hap |
| Dgeo_2186 | Dgeo_2186 | 2305904 | 2306386 | 483 | + | PROTEIN:hypothetical protein |
| Dgeo_2187 | Dgeo_2187 | 2306457 | 2307188 | 732 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2188 | Dgeo_2188 | 2307247 | 2309697 | 2451 | + | PROTEIN:glutaminyl-tRNA synthetase |
| Dgeo_2189 | Dgeo_2189 | 2309706 | 2310524 | 819 | + | PROTEIN:extracellular solute-binding protein |
| Dgeo_2190 | rpsO | 2310648 | 2310923 | 276 | - | PROTEIN:30S ribosomal protein S15 |
| Dgeo_R0055 | Dgeo_R0055 | 2311081 | 2311164 | 84 | + | RNA:Leu tRNA |
| Dgeo_R0056 | ffs | 2311247 | 2311352 | 106 | + | RNA:SRP RNA; RNA component of signal recognition p |
| Dgeo_2191 | Dgeo_2191 | 2311509 | 2312516 | 1008 | - | PROTEIN:transposase, IS4 |
| Dgeo_2192 | Dgeo_2192 | 2312645 | 2313016 | 372 | - | PROTEIN:hypothetical protein |
| Dgeo_2193 | Dgeo_2193 | 2313068 | 2313787 | 720 | + | PROTEIN:carbonic anhydrase |
| Dgeo_2194 | Dgeo_2194 | 2314038 | 2314772 | 735 | + | PROTEIN:hypothetical protein |
| Dgeo_2195 | Dgeo_2195 | 2315022 | 2315906 | 885 | + | PROTEIN:periplasmic binding protein |
| Dgeo_2196 | Dgeo_2196 | 2315974 | 2316462 | 489 | + | PROTEIN:antibiotic biosynthesis monooxygenase |
| Dgeo_2197 | Dgeo_2197 | 2316335 | 2317342 | 1008 | - | PROTEIN:transposase, IS4 |
| Dgeo_2198 | Dgeo_2198 | 2317532 | 2319037 | 1506 | - | PROTEIN:amidohydrolase 3 |
| Dgeo_2199 | Dgeo_2199 | 2319122 | 2320156 | 1035 | + | PROTEIN:response regulator receiver/SARP domain-co |
| Dgeo_2200 | Dgeo_2200 | 2320255 | 2320800 | 546 | + | PROTEIN:hypothetical protein |
| Dgeo_2201 | Dgeo_2201 | 2320945 | 2323602 | 2658 | + | PROTEIN:alanyl-tRNA synthetase |
| Dgeo_2202 | Dgeo_2202 | 2323894 | 2324370 | 477 | - | PROTEIN:MerR family transcriptional regulator |
| Dgeo_2203 | Dgeo_2203 | 2324380 | 2325486 | 1107 | - | PROTEIN:major facilitator transporter |
| Dgeo_2204 | Dgeo_2204 | 2325860 | 2326255 | 396 | - | PROTEIN:hypothetical protein |
| Dgeo_2205 | Dgeo_2205 | 2326349 | 2327440 | 1092 | + | PROTEIN:transposase, IS4 |
| Dgeo_2206 | Dgeo_2206 | 2327542 | 2328402 | 861 | - | PROTEIN:thiosulfate sulfurtransferase |
| Dgeo_2207 | Dgeo_2207 | 2328518 | 2328988 | 471 | + | PROTEIN:Fe-S metabolism associated SufE |
| Dgeo_2208 | Dgeo_2208 | 2328969 | 2329766 | 798 | - | PROTEIN:transposase, IS4 |
| Dgeo_2209 | Dgeo_2209 | 2329902 | 2330501 | 600 | - | PROTEIN:Ham1-like protein protein |
| Dgeo_2210 | Dgeo_2210 | 2330575 | 2331276 | 702 | - | PROTEIN:hypothetical protein |
| Dgeo_2211 | Dgeo_2211 | 2331358 | 2332290 | 933 | - | PROTEIN:Mg2+ transporter protein, CorA-like protei |
| Dgeo_2212 | Dgeo_2212 | 2332553 | 2332789 | 237 | - | PROTEIN:4Fe-4S ferredoxin, iron-sulfur binding |
| Dgeo_2213 | Dgeo_2213 | 2332892 | 2333686 | 795 | - | PROTEIN:hypothetical protein |
| Dgeo_2214 | Dgeo_2214 | 2333683 | 2335125 | 1443 | - | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_2215 | Dgeo_2215 | 2335122 | 2335784 | 663 | - | PROTEIN:two component transcriptional regulator |
| Dgeo_2216 | Dgeo_2216 | 2336208 | 2336507 | 300 | + | PROTEIN:ribosomal protein L21 |
| Dgeo_2217 | rpmA | 2336523 | 2336798 | 276 | + | PROTEIN:50S ribosomal protein L27 |
| Dgeo_2218 | obgE | 2336992 | 2338293 | 1302 | + | PROTEIN:GTPase ObgE |
| Dgeo_2219 | Dgeo_2219 | 2338322 | 2339086 | 765 | - | PROTEIN:peptidylprolyl isomerase |
| Dgeo_2220 | Dgeo_2220 | 2339152 | 2339781 | 630 | + | PROTEIN:cytidylate kinase |
| Dgeo_2221 | Dgeo_2221 | 2339789 | 2340385 | 597 | + | PROTEIN:phospholipid/glycerol acyltransferase |
| Dgeo_2222 | Dgeo_2222 | 2340452 | 2343127 | 2676 | - | PROTEIN:peptidase S8 and S53, subtilisin, kexin, s |
| Dgeo_2223 | Dgeo_2223 | 2343146 | 2344177 | 1032 | - | PROTEIN:peptidase-like protein |
| Dgeo_2224 | Dgeo_2224 | 2344401 | 2345627 | 1227 | - | PROTEIN:peptidase M16-like protein |
| Dgeo_2225 | Dgeo_2225 | 2345624 | 2346886 | 1263 | - | PROTEIN:peptidase M16-like protein |
| Dgeo_2226 | Dgeo_2226 | 2346930 | 2347835 | 906 | - | PROTEIN:pseudouridine synthase, RluD |
| Dgeo_2227 | Dgeo_2227 | 2347886 | 2348404 | 519 | + | PROTEIN:peptidase M23B |
| Dgeo_2228 | Dgeo_2228 | 2348373 | 2349056 | 684 | - | PROTEIN:phosphoglycerate mutase |
| Dgeo_2229 | Dgeo_2229 | 2349085 | 2349690 | 606 | - | PROTEIN:diguanylate cyclase |
| Dgeo_2230 | groES | 2349965 | 2350252 | 288 | + | PROTEIN:co-chaperonin GroES |
| Dgeo_2231 | groEL | 2350311 | 2351948 | 1638 | + | PROTEIN:chaperonin GroEL |
| Dgeo_2232 | Dgeo_2232 | 2352130 | 2353356 | 1227 | - | PROTEIN:cofactor-independent phosphoglycerate muta |
| Dgeo_R0057 | Dgeo_R0057 | 2353553 | 2353629 | 77 | - | RNA:Arg tRNA |
| Dgeo_R0058 | Dgeo_R0058 | 2353665 | 2353738 | 74 | - | RNA:Gly tRNA |
| Dgeo_R0059 | Dgeo_R0059 | 2353766 | 2353842 | 77 | - | RNA:Pro tRNA |
| Dgeo_2233 | Dgeo_2233 | 2354095 | 2354850 | 756 | - | PROTEIN:peptidoglycan binding domain-containing pr |
| Dgeo_2234 | Dgeo_2234 | 2354942 | 2355592 | 651 | + | PROTEIN:tRNA (guanosine-2'-O-)-methyltransferase |
| Dgeo_2235 | Dgeo_2235 | 2355713 | 2356537 | 825 | + | PROTEIN:integral membrane protein TerC |
| Dgeo_2236 | Dgeo_2236 | 2356781 | 2357599 | 819 | - | PROTEIN:AraC family transcriptional regulator |
| Dgeo_2237 | Dgeo_2237 | 2357628 | 2357987 | 360 | - | PROTEIN:hypothetical protein |
| Dgeo_2238 | Dgeo_2238 | 2358053 | 2359006 | 954 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_2239 | Dgeo_2239 | 2359272 | 2360237 | 966 | + | PROTEIN:aldo/keto reductase |
| Dgeo_R0060 | Dgeo_R0060 | 2360347 | 2361839 | 1493 | - | RNA:16S ribosomal RNA |
| Dgeo_2240 | Dgeo_2240 | 2362180 | 2364045 | 1866 | - | PROTEIN:carboxylyase-like protein |
| Dgeo_2241 | Dgeo_2241 | 2364362 | 2365564 | 1203 | + | PROTEIN:peptidase M24 |
| Dgeo_2242 | Dgeo_2242 | 2365653 | 2366657 | 1005 | - | PROTEIN:UDP-glucose 4-epimerase |
| Dgeo_2243 | Dgeo_2243 | 2366707 | 2367726 | 1020 | - | PROTEIN:LacI family transcription regulator |
| Dgeo_2244 | Dgeo_2244 | 2367808 | 2369010 | 1203 | + | PROTEIN:major facilitator transporter |
| Dgeo_2245 | Dgeo_2245 | 2369011 | 2370219 | 1209 | - | PROTEIN:citrate synthase |
| Dgeo_2246 | Dgeo_2246 | 2370288 | 2371598 | 1311 | + | PROTEIN:pyruvate carboxyltransferase |
| Dgeo_2247 | Dgeo_2247 | 2371623 | 2372423 | 801 | + | PROTEIN:hypothetical protein |
| Dgeo_R0061 | Dgeo_R0061 | 2372571 | 2372646 | 76 | - | RNA:Gly tRNA |
| Dgeo_R0062 | Dgeo_R0062 | 2372657 | 2372773 | 117 | - | RNA:5S ribosomal RNA |
| Dgeo_R0063 | Dgeo_R0063 | 2372857 | 2375746 | 2890 | - | RNA:23S ribosomal RNA |
| Dgeo_2248 | Dgeo_2248 | 2375989 | 2376456 | 468 | - | PROTEIN:S-ribosylhomocysteinase |
| Dgeo_2249 | Dgeo_2249 | 2376455 | 2377042 | 588 | + | PROTEIN:lipoprotein signal peptidase |
| Dgeo_2250 | Dgeo_2250 | 2377104 | 2377319 | 216 | - | PROTEIN:ribosomal protein L28 |
| Dgeo_2251 | Dgeo_2251 | 2377546 | 2377815 | 270 | - | PROTEIN:transglycosylase-associated protein |
| Dgeo_2252 | Dgeo_2252 | 2378110 | 2379147 | 1038 | + | PROTEIN:threonine synthase |
| Dgeo_2253 | Dgeo_2253 | 2379213 | 2380166 | 954 | + | PROTEIN:homoserine kinase |
| Dgeo_2254 | Dgeo_2254 | 2380122 | 2380433 | 312 | - | PROTEIN:hypothetical protein |
| Dgeo_2255 | Dgeo_2255 | 2380553 | 2382790 | 2238 | - | PROTEIN:MMPL |
| Dgeo_2256 | Dgeo_2256 | 2382980 | 2383759 | 780 | - | PROTEIN:aminoglycoside phosphotransferase |
| Dgeo_2257 | Dgeo_2257 | 2383840 | 2384745 | 906 | + | PROTEIN:hypothetical protein |
| Dgeo_2258 | Dgeo_2258 | 2384742 | 2385116 | 375 | + | PROTEIN:hypothetical protein |
| Dgeo_2259 | Dgeo_2259 | 2385125 | 2385919 | 795 | + | PROTEIN:CDP-alcohol phosphatidyltransferase |
| Dgeo_2260 | Dgeo_2260 | 2385933 | 2386607 | 675 | - | PROTEIN:hypothetical protein |
| Dgeo_2261 | Dgeo_2261 | 2386668 | 2387327 | 660 | - | PROTEIN:beta-lactamase-like protein |
| Dgeo_2262 | Dgeo_2262 | 2387367 | 2388317 | 951 | - | PROTEIN:DNA polymerase III subunit, putative |
| Dgeo_2263 | Dgeo_2263 | 2388417 | 2389169 | 753 | - | PROTEIN:metallophosphoesterase |
| Dgeo_2264 | Dgeo_2264 | 2389673 | 2390593 | 921 | - | PROTEIN:Nitrilase/cyanide hydratase and apolipopro |
| Dgeo_2265 | Dgeo_2265 | 2390644 | 2391579 | 936 | + | PROTEIN:metalloenzyme |
| Dgeo_2266 | Dgeo_2266 | 2391762 | 2392406 | 645 | - | PROTEIN:phosphoribosylanthranilate isomerase |
| Dgeo_2267 | Dgeo_2267 | 2392403 | 2392639 | 237 | - | PROTEIN:hypothetical protein |
| Dgeo_2268 | Dgeo_2268 | 2392709 | 2393374 | 666 | - | PROTEIN:DtxR family iron dependent repressor |
| Dgeo_2269 | Dgeo_2269 | 2393448 | 2394032 | 585 | + | PROTEIN:ECF subfamily RNA polymerase sigma-24 fact |
| Dgeo_2270 | Dgeo_2270 | 2394136 | 2394411 | 276 | + | PROTEIN:hypothetical protein |
| Dgeo_2271 | Dgeo_2271 | 2394408 | 2395202 | 795 | + | PROTEIN:putative sigma E regulatory protein, MucB/ |
| Dgeo_2272 | Dgeo_2272 | 2395236 | 2396411 | 1176 | - | PROTEIN:putative transposase, IS891/IS1136/IS1341 |
| Dgeo_2273 | Dgeo_2273 | 2396412 | 2396810 | 399 | - | PROTEIN:transposase IS200-like protein |
| Dgeo_2274 | Dgeo_2274 | 2397073 | 2397654 | 582 | + | PROTEIN:hypothetical protein |
| Dgeo_2275 | Dgeo_2275 | 2397824 | 2398624 | 801 | + | PROTEIN:hypothetical protein |
| Dgeo_2276 | Dgeo_2276 | 2398966 | 2399763 | 798 | + | PROTEIN:transposase, IS4 |
| Dgeo_2277 | Dgeo_2277 | 2399779 | 2400711 | 933 | - | PROTEIN:phage integrase |
| Dgeo_2278 | Dgeo_2278 | 2400708 | 2400941 | 234 | - | PROTEIN:hypothetical protein |
| Dgeo_2279 | Dgeo_2279 | 2401336 | 2402271 | 936 | + | PROTEIN:hypothetical protein |
| Dgeo_2280 | Dgeo_2280 | 2402370 | 2403545 | 1176 | + | PROTEIN:twin-arginine translocation pathway signal |
| Dgeo_2281 | Dgeo_2281 | 2403545 | 2403985 | 441 | + | PROTEIN:hypothetical protein |
| Dgeo_2282 | Dgeo_2282 | 2404292 | 2404666 | 375 | + | PROTEIN:hypothetical protein |
| Dgeo_2283 | Dgeo_2283 | 2404801 | 2406804 | 2004 | + | PROTEIN:transketolase |
| Dgeo_2284 | Dgeo_2284 | 2406815 | 2407543 | 729 | - | PROTEIN:guanylate kinase |
| Dgeo_2285 | Dgeo_2285 | 2407618 | 2408073 | 456 | - | PROTEIN:hypothetical protein |
| Dgeo_2286 | Dgeo_2286 | 2408231 | 2408524 | 294 | - | PROTEIN:AsnC family transcriptional regulator |
| Dgeo_2287 | Dgeo_2287 | 2408672 | 2409727 | 1056 | + | PROTEIN:AsnC family transcriptional regulator |
| Dgeo_2288 | Dgeo_2288 | 2409786 | 2410706 | 921 | - | PROTEIN:short-chain dehydrogenase/reductase SDR |
| Dgeo_2289 | Dgeo_2289 | 2410801 | 2411316 | 516 | + | PROTEIN:hypothetical protein |
| Dgeo_2290 | Dgeo_2290 | 2411309 | 2412037 | 729 | + | PROTEIN:HhH-GPD |
| Dgeo_2291 | Dgeo_2291 | 2412059 | 2413018 | 960 | - | PROTEIN:methionyl-tRNA formyltransferase |
| Dgeo_2292 | Dgeo_2292 | 2413015 | 2413665 | 651 | - | PROTEIN:peptide deformylase |
| Dgeo_2293 | Dgeo_2293 | 2413822 | 2414976 | 1155 | + | PROTEIN:tetratricopeptide TPR_2 |
| Dgeo_2294 | Dgeo_2294 | 2415034 | 2415576 | 543 | + | PROTEIN:hypothetical protein |
| Dgeo_2295 | Dgeo_2295 | 2415498 | 2416079 | 582 | - | PROTEIN:hypothetical protein |
| Dgeo_2296 | Dgeo_2296 | 2416258 | 2417397 | 1140 | - | PROTEIN:alcohol dehydrogenase GroES-like protein |
| Dgeo_2297 | Dgeo_2297 | 2417475 | 2417960 | 486 | - | PROTEIN:hypothetical protein |
| Dgeo_2299 | Dgeo_2299 | 2418765 | 2418989 | 225 | - | PROTEIN:hypothetical protein |
| Dgeo_2300 | Dgeo_2300 | 2419007 | 2419318 | 312 | - | PROTEIN:hypothetical protein |
| Dgeo_2301 | Dgeo_2301 | 2419744 | 2420589 | 846 | + | PROTEIN:hypothetical protein |
| Dgeo_2303 | Dgeo_2303 | 2422129 | 2422497 | 369 | - | PROTEIN:hypothetical protein |
| Dgeo_2304 | Dgeo_2304 | 2422606 | 2423793 | 1188 | + | PROTEIN:major facilitator transporter |
| Dgeo_2305 | Dgeo_2305 | 2423849 | 2424595 | 747 | - | PROTEIN:LmbE-like protein protein |
| Dgeo_2306 | Dgeo_2306 | 2424592 | 2426076 | 1485 | - | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_2307 | Dgeo_2307 | 2426073 | 2427155 | 1083 | - | PROTEIN:glycosyl transferase family protein |
| Dgeo_2308 | Dgeo_2308 | 2427152 | 2427808 | 657 | - | PROTEIN:phospholipid/glycerol acyltransferase |
| Dgeo_2309 | Dgeo_2309 | 2427805 | 2428728 | 924 | - | PROTEIN:hypothetical protein |
| Dgeo_2310 | Dgeo_2310 | 2428786 | 2430330 | 1545 | - | PROTEIN:FAD dependent oxidoreductase |
| Dgeo_2311 | Dgeo_2311 | 2430396 | 2430584 | 189 | + | PROTEIN:hypothetical protein |
| Dgeo_2312 | Dgeo_2312 | 2430750 | 2431973 | 1224 | + | PROTEIN:hypothetical protein |
| Dgeo_2313 | Dgeo_2313 | 2432029 | 2433033 | 1005 | - | PROTEIN:GCN5-related N-acetyltransferase |
| Dgeo_2314 | Dgeo_2314 | 2433137 | 2434849 | 1713 | - | PROTEIN:peptidase M3A and M3B, thimet/oligopeptida |
| Dgeo_2315 | Dgeo_2315 | 2434984 | 2436162 | 1179 | + | PROTEIN:phosphopantothenoylcysteine decarboxylase/ |
| Dgeo_2316 | Dgeo_2316 | 2436268 | 2436729 | 462 | + | PROTEIN:arginine repressor, ArgR |
| Dgeo_2317 | Dgeo_2317 | 2436787 | 2438361 | 1575 | - | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_2318 | Dgeo_2318 | 2438439 | 2439116 | 678 | - | PROTEIN:two component transcriptional regulator |
| Dgeo_2319 | Dgeo_2319 | 2439173 | 2439754 | 582 | - | PROTEIN:putative adenylate/guanylate cyclase |
| Dgeo_2320 | Dgeo_2320 | 2439840 | 2440277 | 438 | - | PROTEIN:hypothetical protein |
| Dgeo_2321 | Dgeo_2321 | 2440314 | 2440610 | 297 | - | PROTEIN:hypothetical protein |
| Dgeo_2322 | Dgeo_2322 | 2440592 | 2441551 | 960 | - | PROTEIN:4-hydroxybenzoate polyprenyltransferase, p |
| Dgeo_R0064 | Dgeo_R0064 | 2441584 | 2441660 | 77 | - | RNA:Arg tRNA |
| Dgeo_2323 | Dgeo_2323 | 2441754 | 2443157 | 1404 | - | PROTEIN:dihydrolipoamide dehydrogenase |
| Dgeo_2324 | Dgeo_2324 | 2443249 | 2444460 | 1212 | - | PROTEIN:putative oxygen-independent coproporphyrin |
| Dgeo_2325 | Dgeo_2325 | 2444457 | 2445035 | 579 | - | PROTEIN:hypothetical protein |
| Dgeo_2326 | Dgeo_2326 | 2445208 | 2446002 | 795 | - | PROTEIN:hypothetical protein |
| Dgeo_2327 | Dgeo_2327 | 2446024 | 2447190 | 1167 | - | PROTEIN:hypothetical protein |
| Dgeo_2328 | fumC | 2447539 | 2448978 | 1440 | - | PROTEIN:fumarate hydratase |
| Dgeo_2329 | Dgeo_2329 | 2449073 | 2450005 | 933 | + | PROTEIN:AraC family transcriptional regulator |
| Dgeo_2330 | Dgeo_2330 | 2450063 | 2451298 | 1236 | + | PROTEIN:major facilitator transporter |
| Dgeo_2331 | Dgeo_2331 | 2451348 | 2452364 | 1017 | + | PROTEIN:HI0933-like protein protein |
| Dgeo_2332 | Dgeo_2332 | 2452409 | 2453761 | 1353 | + | PROTEIN:VanW |
| Dgeo_2333 | Dgeo_2333 | 2453842 | 2454687 | 846 | - | PROTEIN:parB-like protein partition proteins |
| Dgeo_2334 | Dgeo_2334 | 2454671 | 2455420 | 750 | - | PROTEIN:cobyrinic acid a,c-diamide synthase |
| Dgeo_2335 | Dgeo_2335 | 2455417 | 2456154 | 738 | - | PROTEIN:methyltransferase GidB |
| Dgeo_2336 | Dgeo_2336 | 2456151 | 2456477 | 327 | - | PROTEIN:hypothetical protein |
| Dgeo_2337 | gidA | 2456575 | 2458419 | 1845 | - | PROTEIN:tRNA uridine 5-carboxymethylaminomethyl mo |
| Dgeo_2338 | Dgeo_2338 | 2458567 | 2459676 | 1110 | + | PROTEIN:pyruvate dehydrogenase (lipoamide) |
| Dgeo_2339 | Dgeo_2339 | 2459673 | 2460695 | 1023 | + | PROTEIN:transketolase, central region |
| Dgeo_2340 | Dgeo_2340 | 2460702 | 2460935 | 234 | + | PROTEIN:hypothetical protein |
| Dgeo_2341 | Dgeo_2341 | 2460963 | 2462513 | 1551 | + | PROTEIN:branched-chain alpha-keto acid dehydrogena |
| Dgeo_2342 | Dgeo_2342 | 2462676 | 2463119 | 444 | + | PROTEIN:hypothetical protein |
| Dgeo_2343 | Dgeo_2343 | 2463120 | 2463779 | 660 | + | PROTEIN:two component transcriptional regulator |
| Dgeo_2344 | Dgeo_2344 | 2463873 | 2465234 | 1362 | + | PROTEIN:periplasmic sensor signal transduction his |
| Dgeo_2345 | Dgeo_2345 | 2465263 | 2466339 | 1077 | + | PROTEIN:DNA processing protein DprA, putative |
| Dgeo_2346 | Dgeo_2346 | 2466348 | 2466662 | 315 | - | PROTEIN:hypothetical protein |
| Dgeo_2347 | Dgeo_2347 | 2466734 | 2467021 | 288 | - | PROTEIN:hypothetical protein |