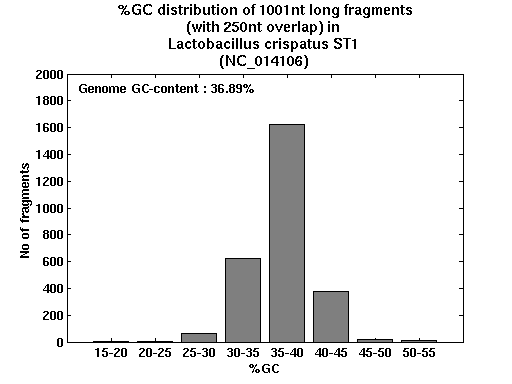
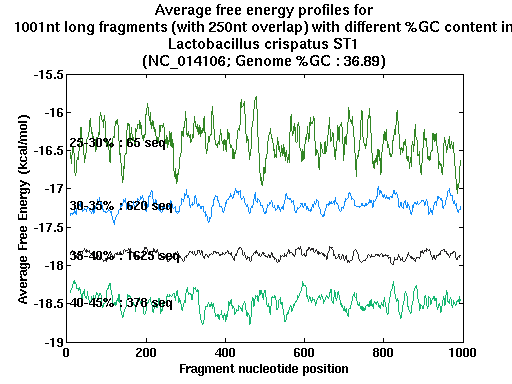
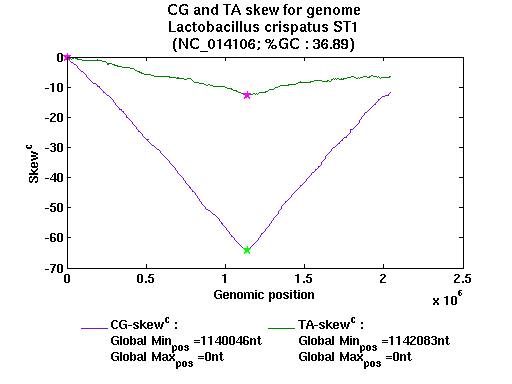
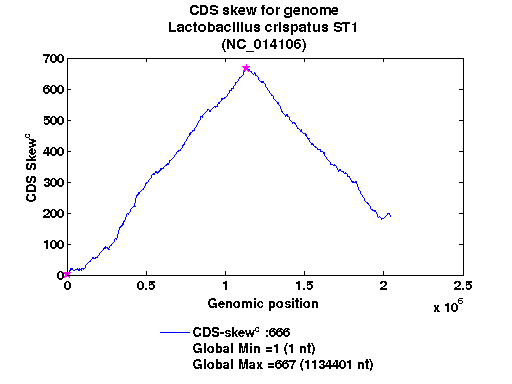
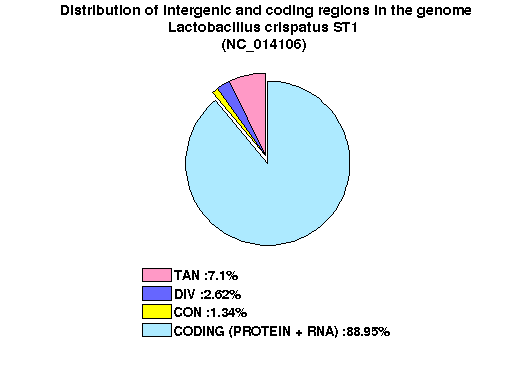
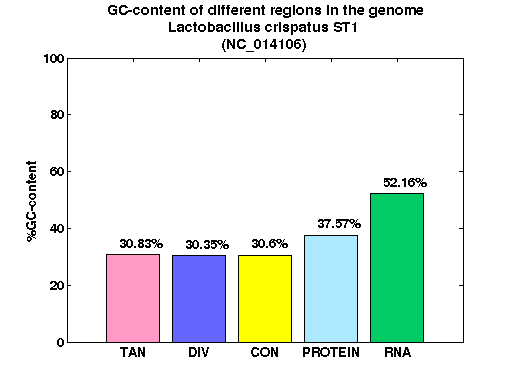
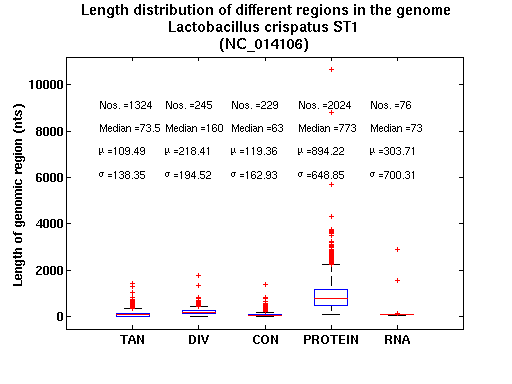
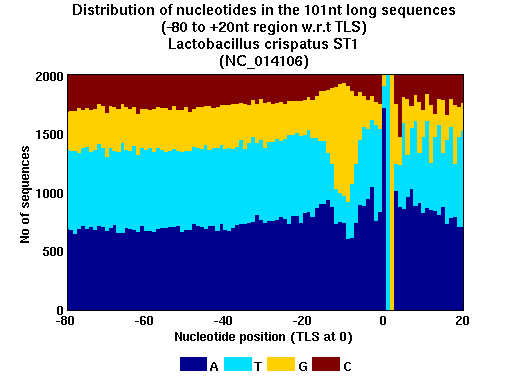
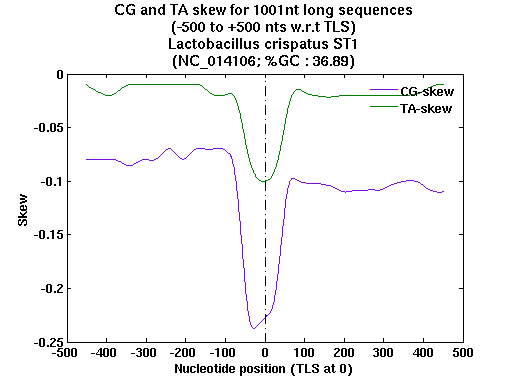
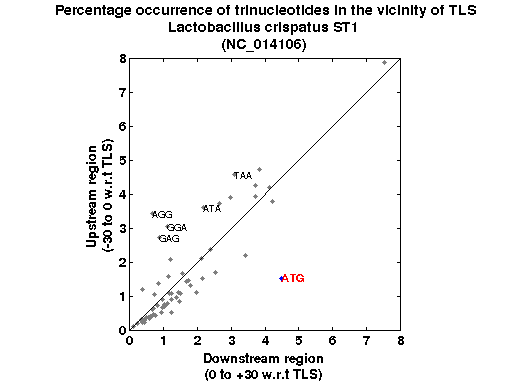
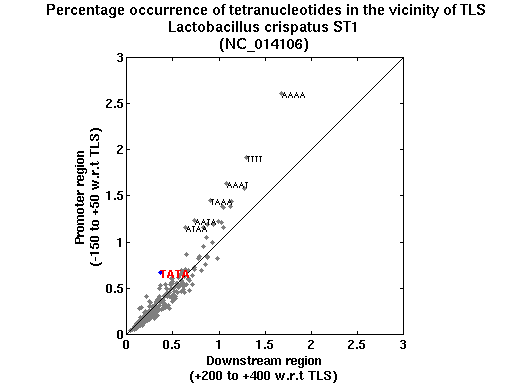
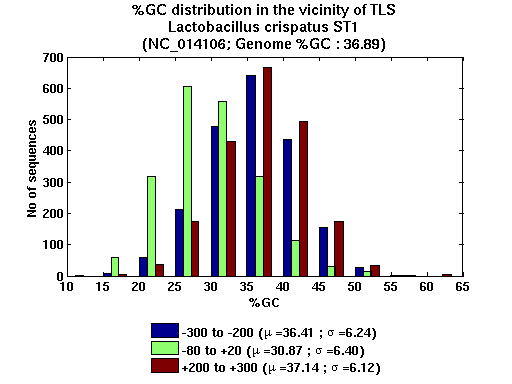
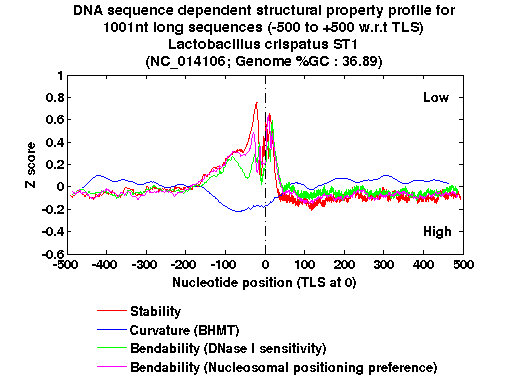
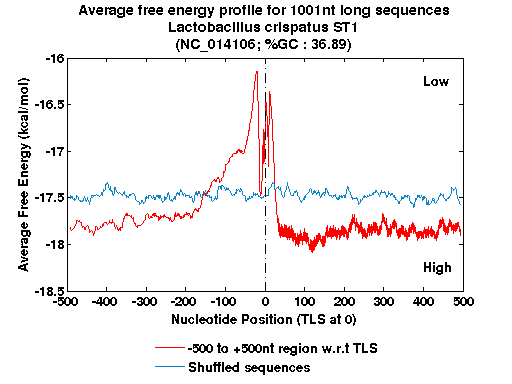
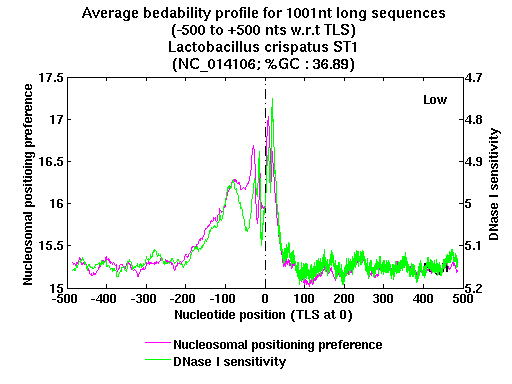
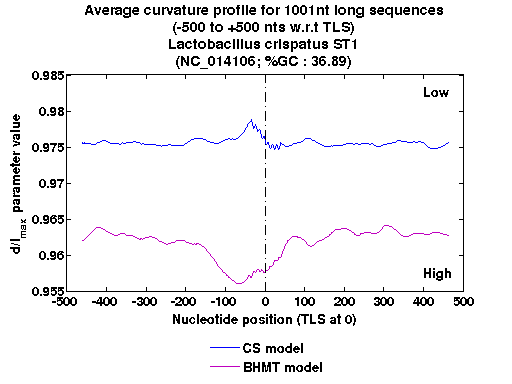
| Gene ID |
Gene name |
Left end
position |
Right end
position |
Length |
Transcription
direction |
Gene Product |
| LCRIS_00001 | dnaA | 1 | 1368 | 1368 | + | PROTEIN:chromosomal replication initiator protein |
| LCRIS_00002 | dnaN | 1543 | 2673 | 1131 | + | PROTEIN:DNA polymerase iii, beta subunit |
| LCRIS_00003 | LCRIS_00003 | 2890 | 3111 | 222 | + | PROTEIN:RNA-binding s4 domain protein |
| LCRIS_00004 | recF | 3120 | 4247 | 1128 | + | PROTEIN:DNA replication and repair protein recf |
| LCRIS_00005 | gyrB | 4248 | 6212 | 1965 | + | PROTEIN:DNA gyrase, b subunit |
| LCRIS_00006 | gyrA | 6222 | 8702 | 2481 | + | PROTEIN:DNA gyrase, a subunit |
| LCRIS_00007 | rpsF | 8920 | 9216 | 297 | + | PROTEIN:30S ribosomal protein s6 |
| LCRIS_00008 | ssb | 9266 | 9784 | 519 | + | PROTEIN:single-stranded DNA-binding protein |
| LCRIS_00009 | rpsR | 9812 | 10048 | 237 | + | PROTEIN:30S ribosomal protein s18 |
| LCRIS_00010 | LCRIS_00010 | 10107 | 10577 | 471 | - | PROTEIN:prophage repressor |
| LCRIS_00011 | LCRIS_00011 | 10682 | 11560 | 879 | + | PROTEIN:bacteriocin helveticin-j |
| LCRIS_00012 | LCRIS_00012 | 11701 | 12807 | 1107 | + | PROTEIN:s-layer protein |
| LCRIS_00013 | LCRIS_00013 | 13028 | 15049 | 2022 | + | PROTEIN:phosphoesterase, dhh family protein |
| LCRIS_00014 | rplI | 15061 | 15516 | 456 | + | PROTEIN:50S ribosomal protein l9 |
| LCRIS_00015 | dnaB1 | 15536 | 16927 | 1392 | + | PROTEIN:replicative DNA helicase |
| LCRIS_00016 | phnD1 | 17095 | 18030 | 936 | + | PROTEIN:phosphonate ABC transporter, phosphonate-b |
| LCRIS_00017 | degV1 | 18140 | 19069 | 930 | + | PROTEIN:degv family protein |
| LCRIS_00018 | scrK | 19185 | 20063 | 879 | + | PROTEIN:fructokinase |
| LCRIS_00019 | LCRIS_00019 | 20211 | 20402 | 192 | + | PROTEIN:hypothetical protein |
| LCRIS_00020 | LCRIS_00020 | 20453 | 20710 | 258 | + | PROTEIN:integral hypothetical protein |
| LCRIS_00021 | LCRIS_00021 | 20762 | 21991 | 1230 | - | PROTEIN:hypothetical protein |
| LCRIS_00022 | mgtC | 22089 | 22790 | 702 | - | PROTEIN:Mg+2 transporter-c (mgtc) family protein |
| LCRIS_02037 | tRNA-Thr | 22965 | 23037 | 73 | + | RNA:Thr tRNA |
| LCRIS_00023 | LCRIS_00023 | 23214 | 23435 | 222 | + | PROTEIN:hypothetical protein |
| LCRIS_00024 | LCRIS_00024 | 23595 | 24662 | 1068 | + | PROTEIN:hypothetical protein |
| LCRIS_00025 | LCRIS_00025 | 24761 | 25534 | 774 | + | PROTEIN:hypothetical protein |
| LCRIS_00027 | LCRIS_00027 | 25602 | 25766 | 165 | - | PROTEIN:adenosylcobalamin biosynthesis protein |
| LCRIS_00026 | LCRIS_00026 | 25759 | 26073 | 315 | + | PROTEIN:lpxtg-motif cell wall anchor |
| LCRIS_00028 | LCRIS_00028 | 26168 | 26428 | 261 | + | PROTEIN:hypothetical protein |
| LCRIS_00029 | LCRIS_00029 | 26758 | 30456 | 3699 | + | PROTEIN:mucus-binding protein |
| LCRIS_00030 | LCRIS_00030 | 30609 | 30776 | 168 | - | PROTEIN:hypothetical protein |
| LCRIS_00031 | LCRIS_00031 | 30842 | 31225 | 384 | - | PROTEIN:transposase |
| LCRIS_00032 | LCRIS_00032 | 31185 | 31433 | 249 | - | PROTEIN:hypothetical protein |
| LCRIS_00033 | oppA1 | 31856 | 33616 | 1761 | - | PROTEIN:oligopeptide ABC transporter, oligopeptide |
| LCRIS_00034 | LCRIS_00034 | 33813 | 34847 | 1035 | + | PROTEIN:permease |
| LCRIS_00035 | LCRIS_00035 | 34847 | 35419 | 573 | + | PROTEIN:esterase |
| LCRIS_00036 | LCRIS_00036 | 35453 | 35635 | 183 | - | PROTEIN:hypothetical protein |
| LCRIS_00037 | LCRIS_00037 | 35819 | 37153 | 1335 | + | PROTEIN:sugar transporter |
| LCRIS_00038 | LCRIS_00038 | 37561 | 38322 | 762 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00039 | LCRIS_00039 | 38319 | 39215 | 897 | - | PROTEIN:ABC transporter, permease protein |
| LCRIS_00040 | LCRIS_00040 | 39212 | 40204 | 993 | - | PROTEIN:ABC transporter, substrate-binding protein |
| LCRIS_00041 | LCRIS_00041 | 40487 | 42094 | 1608 | - | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_00042 | LCRIS_00042 | 42184 | 42996 | 813 | - | PROTEIN:transcriptional regulator, xre family |
| LCRIS_00043 | LCRIS_00043 | 43109 | 43816 | 708 | + | PROTEIN:had superfamily hydrolase |
| LCRIS_00044 | LCRIS_00044 | 43813 | 44682 | 870 | - | PROTEIN:Mg+2 and co2 transporter |
| LCRIS_00045 | LCRIS_00045 | 44798 | 45655 | 858 | + | PROTEIN:oxidoreductase, aldo/keto reductase family |
| LCRIS_00046 | LCRIS_00046 | 45722 | 46426 | 705 | - | PROTEIN:hypothetical protein |
| LCRIS_00047 | LCRIS_00047 | 46846 | 47664 | 819 | - | PROTEIN:transcriptional regulator |
| LCRIS_00048 | LCRIS_00048 | 47721 | 47891 | 171 | - | PROTEIN:hypothetical protein |
| LCRIS_00049 | LCRIS_00049 | 48029 | 49954 | 1926 | + | PROTEIN:translation elongation factor, homologous |
| LCRIS_00050 | LCRIS_00050 | 50123 | 50941 | 819 | + | PROTEIN:had superfamily hydrolase |
| LCRIS_00051 | ldhA | 51010 | 52023 | 1014 | - | PROTEIN:d-lactate dehydrogenase |
| LCRIS_00052 | LCRIS_00052 | 52381 | 52632 | 252 | + | PROTEIN:hypothetical protein |
| LCRIS_00053 | LCRIS_00053 | 52592 | 53131 | 540 | + | PROTEIN:lpxtg-motif cell wall anchor |
| LCRIS_00054 | xthA | 53686 | 54513 | 828 | + | PROTEIN:exodeoxyribonuclease iii |
| LCRIS_00055 | LCRIS_00055 | 54587 | 56029 | 1443 | + | PROTEIN:glutamate/gamma-aminobutyrate antiporter |
| LCRIS_00056 | LCRIS_00056 | 56095 | 58923 | 2829 | + | PROTEIN:DNA/RNA helicase, dead/deah box family |
| LCRIS_00057 | LCRIS_00057 | 58958 | 60010 | 1053 | - | PROTEIN:penicillin-binding protein |
| LCRIS_00058 | LCRIS_00058 | 60241 | 61431 | 1191 | - | PROTEIN:hypothetical protein |
| LCRIS_00059 | def1 | 61493 | 61906 | 414 | + | PROTEIN:peptide deformylase |
| LCRIS_00060 | LCRIS_00060 | 62484 | 62576 | 93 | - | PROTEIN:hypothetical protein |
| LCRIS_02033 | 16srRNA | 62524 | 64075 | 1552 | + | RNA:16S ribosomal RNA |
| LCRIS_00061 | LCRIS_00061 | 62786 | 63220 | 435 | - | PROTEIN:pg1 protein, homology to Homo sapiens |
| LCRIS_00062 | LCRIS_00062 | 63654 | 64007 | 354 | + | PROTEIN:hypothetical protein |
| LCRIS_02038 | tRNA-Ile | 64212 | 64286 | 75 | + | RNA:Ile tRNA |
| LCRIS_02039 | tRNA-Ala | 64343 | 64415 | 73 | + | RNA:Ala tRNA |
| LCRIS_02029 | 23srRNA | 64538 | 67443 | 2906 | + | RNA:23S ribosomal RNA |
| LCRIS_00063 | LCRIS_00063 | 65092 | 65562 | 471 | - | PROTEIN:hypothetical protein |
| LCRIS_00064 | LCRIS_00064 | 66983 | 67300 | 318 | - | PROTEIN:cell wall-associated hydrolase |
| LCRIS_02025 | 5srRNA | 67516 | 67631 | 116 | + | RNA:5S ribosomal RNA |
| LCRIS_02040 | tRNA-Asn | 67644 | 67719 | 76 | + | RNA:Asn tRNA |
| LCRIS_00065 | LCRIS_00065 | 68238 | 68672 | 435 | + | PROTEIN:acetyltransferase |
| LCRIS_00066 | LCRIS_00066 | 68891 | 69112 | 222 | - | PROTEIN:steroid binding protein |
| LCRIS_00067 | arbZ | 69205 | 70074 | 870 | - | PROTEIN:phospho-beta-glycosidase |
| LCRIS_00068 | arbY | 70071 | 71018 | 948 | - | PROTEIN:lipopolysaccharide biosynthesis glycosyltr |
| LCRIS_00069 | LCRIS_00069 | 71032 | 71985 | 954 | - | PROTEIN:lipopolysaccharide biosynthesis glycosyltr |
| LCRIS_00070 | arbx | 72003 | 72839 | 837 | - | PROTEIN:lipopolysaccharide biosynthesis glycosyltr |
| LCRIS_00071 | arbV | 72856 | 73617 | 762 | - | PROTEIN:1-acyl-sn-glycerol-3-phosphate acyltransfe |
| LCRIS_02100 | tRNA-Lys | 73797 | 73869 | 73 | - | RNA:Lys tRNA |
| LCRIS_00072 | LCRIS_00072 | 74002 | 74223 | 222 | - | PROTEIN:hypothetical protein |
| LCRIS_02099 | tRNA-Lys | 74342 | 74414 | 73 | - | RNA:Lys tRNA |
| LCRIS_00073 | vicR | 74551 | 75267 | 717 | + | PROTEIN:response regulator |
| LCRIS_00074 | vicK | 75338 | 77191 | 1854 | + | PROTEIN:sensor protein |
| LCRIS_00075 | LCRIS_00075 | 77172 | 78533 | 1362 | + | PROTEIN:hypothetical protein |
| LCRIS_00076 | LCRIS_00076 | 78536 | 79360 | 825 | + | PROTEIN:hypothetical protein |
| LCRIS_00077 | LCRIS_00077 | 79373 | 80170 | 798 | + | PROTEIN:metallo-beta-lactamase family protein |
| LCRIS_00078 | htrA | 80232 | 81512 | 1281 | + | PROTEIN:serine protease |
| LCRIS_00079 | ybeA | 82030 | 82509 | 480 | + | PROTEIN:alpha/beta knot family protein |
| LCRIS_00080 | LCRIS_00080 | 83009 | 83554 | 546 | + | PROTEIN:peptidase |
| LCRIS_00081 | pepI | 83578 | 84459 | 882 | - | PROTEIN:proline iminopeptidase |
| LCRIS_00082 | cbiQ1 | 84468 | 85121 | 654 | - | PROTEIN:ABC transporter, permease protein |
| LCRIS_00083 | cbiO1 | 85126 | 86478 | 1353 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00084 | LCRIS_00084 | 86635 | 87576 | 942 | + | PROTEIN:regulatory protein |
| LCRIS_00085 | htpX | 87612 | 88508 | 897 | - | PROTEIN:protease htpx homolog |
| LCRIS_00086 | lemA | 88527 | 89087 | 561 | - | PROTEIN:lema family protein |
| LCRIS_00087 | LCRIS_00087 | 89184 | 90290 | 1107 | - | PROTEIN:cation transporter |
| LCRIS_00088 | LCRIS_00088 | 90490 | 90894 | 405 | + | PROTEIN:hypothetical protein |
| LCRIS_00089 | glpQ1 | 90988 | 92766 | 1779 | + | PROTEIN:glycerophosphodiester phosphodiesterase |
| LCRIS_00090 | LCRIS_00090 | 93133 | 93999 | 867 | + | PROTEIN:hypothetical protein |
| LCRIS_00091 | LCRIS_00091 | 93983 | 94138 | 156 | + | PROTEIN:hypothetical protein |
| LCRIS_00092 | LCRIS_00092 | 94185 | 95438 | 1254 | + | PROTEIN:glycosyltransferase |
| LCRIS_00093 | LCRIS_00093 | 95456 | 96559 | 1104 | + | PROTEIN:endoglucanase |
| LCRIS_00094 | LCRIS_00094 | 96648 | 97292 | 645 | + | PROTEIN:hd superfamily hydrolase |
| LCRIS_00095 | LCRIS_00095 | 97293 | 98210 | 918 | + | PROTEIN:restriction endonuclease |
| LCRIS_00096 | LCRIS_00096 | 98280 | 98750 | 471 | - | PROTEIN:hypothetical protein |
| LCRIS_00097 | LCRIS_00097 | 98832 | 99587 | 756 | - | PROTEIN:amino acid ABC transporter, ATP-binding pr |
| LCRIS_00098 | LCRIS_00098 | 99587 | 101200 | 1614 | - | PROTEIN:amino acid ABC transporter, permease compo |
| LCRIS_00099 | LCRIS_00099 | 101533 | 102165 | 633 | - | PROTEIN:3-methyladenine DNA glycosylase |
| LCRIS_00100 | LCRIS_00100 | 102198 | 102563 | 366 | + | PROTEIN:hypothetical protein |
| LCRIS_00101 | LCRIS_00101 | 102560 | 103348 | 789 | - | PROTEIN:hypothetical protein |
| LCRIS_00102 | ogt1 | 103335 | 103853 | 519 | - | PROTEIN:methylated-DNA--protein-cysteine s-methylt |
| LCRIS_00103 | npdA | 103906 | 104604 | 699 | - | PROTEIN:nad-dependent protein deacetylase, sir2 fa |
| LCRIS_00104 | LCRIS_00104 | 104694 | 105179 | 486 | + | PROTEIN:hypothetical protein |
| LCRIS_00105 | cls1 | 105180 | 106637 | 1458 | - | PROTEIN:cardiolipin synthetase |
| LCRIS_00106 | LCRIS_00106 | 106657 | 107583 | 927 | - | PROTEIN:alpha/beta hydrolase superfamily protein |
| LCRIS_00107 | rnhA | 107679 | 108449 | 771 | + | PROTEIN:ribonuclease h |
| LCRIS_00108 | LCRIS_00108 | 108477 | 109430 | 954 | - | PROTEIN:extracellular protein |
| LCRIS_00109 | LCRIS_00109 | 109591 | 110403 | 813 | + | PROTEIN:polysaccharide biosynthesis protein |
| LCRIS_00110 | LCRIS_00110 | 110412 | 111068 | 657 | + | PROTEIN:oxidoreductase |
| LCRIS_00111 | LCRIS_00111 | 111137 | 111961 | 825 | + | PROTEIN:transcriptional regulator, xre family |
| LCRIS_00112 | LCRIS_00112 | 112158 | 112418 | 261 | + | PROTEIN:hypothetical protein |
| LCRIS_00113 | LCRIS_00113 | 112432 | 112845 | 414 | + | PROTEIN:hypothetical protein |
| LCRIS_00114 | LCRIS_00114 | 113153 | 113908 | 756 | + | PROTEIN:hypothetical protein |
| LCRIS_00115 | LCRIS_00115 | 113911 | 114963 | 1053 | + | PROTEIN:hypothetical protein |
| LCRIS_00116 | LCRIS_00116 | 114986 | 116221 | 1236 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00117 | LCRIS_00117 | 116199 | 116945 | 747 | + | PROTEIN:hypothetical protein |
| LCRIS_00118 | LCRIS_00118 | 117568 | 118746 | 1179 | + | PROTEIN:transposase |
| LCRIS_00119 | LCRIS_00119 | 118830 | 119501 | 672 | + | PROTEIN:hypothetical protein |
| LCRIS_00120 | LCRIS_00120 | 119501 | 119662 | 162 | + | PROTEIN:hypothetical protein |
| LCRIS_00121 | LCRIS_00121 | 119816 | 121222 | 1407 | + | PROTEIN:transposase |
| LCRIS_00122 | prs1 | 121360 | 122346 | 987 | + | PROTEIN:ribose-phosphate pyrophosphokinase |
| LCRIS_00123 | LCRIS_00123 | 122511 | 123194 | 684 | + | PROTEIN:transcriptional regulator |
| LCRIS_00124 | LCRIS_00124 | 123207 | 124031 | 825 | + | PROTEIN:hypothetical protein |
| LCRIS_00125 | glnP1 | 124158 | 125642 | 1485 | + | PROTEIN:amino acid ABC transporter, amino acid-bin |
| LCRIS_00126 | glnQ1 | 125635 | 126372 | 738 | + | PROTEIN:amino acid ABC transporter, ATP-binding pr |
| LCRIS_00127 | LCRIS_00127 | 126408 | 127658 | 1251 | - | PROTEIN:hypothetical protein |
| LCRIS_00128 | ddl | 127811 | 128893 | 1083 | + | PROTEIN:D-alanine--D-alanine ligase |
| LCRIS_00129 | LCRIS_00129 | 128921 | 129670 | 750 | + | PROTEIN:transcriptional regulator |
| LCRIS_00130 | LCRIS_00130 | 129761 | 130894 | 1134 | + | PROTEIN:hypothetical protein |
| LCRIS_00131 | LCRIS_00131 | 130891 | 131676 | 786 | + | PROTEIN:multitranshypothetical protein |
| LCRIS_00132 | LCRIS_00132 | 131648 | 132481 | 834 | + | PROTEIN:esterase/lipase |
| LCRIS_00133 | LCRIS_00133 | 132453 | 132722 | 270 | + | PROTEIN:hypothetical protein |
| LCRIS_00134 | LCRIS_00134 | 132772 | 135780 | 3009 | - | PROTEIN:alpha-glucosidase |
| LCRIS_00135 | nagA | 135934 | 137046 | 1113 | + | PROTEIN:N-acetylglucosamine-6-phosphate deacetylas |
| LCRIS_00136 | LCRIS_00136 | 137281 | 138459 | 1179 | - | PROTEIN:transposase |
| LCRIS_00137 | LCRIS_00137 | 140215 | 140511 | 297 | + | PROTEIN:hypothetical protein |
| LCRIS_00138 | LCRIS_00138 | 140513 | 140866 | 354 | + | PROTEIN:hypothetical protein |
| LCRIS_00139 | LCRIS_00139 | 141088 | 142485 | 1398 | + | PROTEIN:atpase |
| LCRIS_00140 | LCRIS_00140 | 142534 | 142686 | 153 | + | PROTEIN:hypothetical protein |
| LCRIS_00141 | ntd | 142855 | 143331 | 477 | - | PROTEIN:nucleoside deoxyribosyltransferase |
| LCRIS_00142 | LCRIS_00142 | 143446 | 143943 | 498 | + | PROTEIN:PTS system iia component |
| LCRIS_00143 | LCRIS_00143 | 143961 | 144764 | 804 | - | PROTEIN:protein tyrosine phosphatase |
| LCRIS_00144 | usp | 144909 | 145394 | 486 | + | PROTEIN:universal stress protein family |
| LCRIS_00145 | LCRIS_00145 | 145477 | 146511 | 1035 | - | PROTEIN:transposase |
| LCRIS_00146 | phnD2 | 146916 | 147845 | 930 | + | PROTEIN:phosphonate ABC transporter, phosphonate-b |
| LCRIS_00147 | phnC | 147876 | 148649 | 774 | + | PROTEIN:phosphonate ABC transporter, ATP-binding p |
| LCRIS_00148 | phnE1 | 148649 | 149440 | 792 | + | PROTEIN:phosphonate ABC transporter, permease prot |
| LCRIS_00149 | phnE2 | 149440 | 150252 | 813 | + | PROTEIN:phosphonate ABC transporter, permease prot |
| LCRIS_00150 | tag | 150307 | 150876 | 570 | - | PROTEIN:DNA-3-methyladenine glycosylase i |
| LCRIS_00151 | LCRIS_00151 | 151035 | 151520 | 486 | + | PROTEIN:hypothetical protein |
| LCRIS_00152 | LCRIS_00152 | 151577 | 152974 | 1398 | + | PROTEIN:5'-nucleotidase/2',3'-cyclic phosphodieste |
| LCRIS_00153 | asnB | 153098 | 155047 | 1950 | + | PROTEIN:asparagine synthase |
| LCRIS_00154 | LCRIS_00154 | 155069 | 156352 | 1284 | + | PROTEIN:ATP-grasp enzyme |
| LCRIS_00155 | nrdD | 156526 | 158733 | 2208 | + | PROTEIN:anaerobic ribonucleoside-triphosphate redu |
| LCRIS_00156 | nrdG | 158733 | 159446 | 714 | + | PROTEIN:anaerobic ribonucleoside-triphosphate redu |
| LCRIS_00157 | LCRIS_00157 | 159498 | 160076 | 579 | + | PROTEIN:metal-sulfur cluster biosynthetic enzyme |
| LCRIS_00158 | LCRIS_00158 | 160069 | 161265 | 1197 | + | PROTEIN:hypothetical protein |
| LCRIS_00159 | LCRIS_00159 | 161382 | 162599 | 1218 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00160 | pepO1 | 162681 | 164627 | 1947 | + | PROTEIN:endopeptidase o |
| LCRIS_00161 | kup1 | 164796 | 166817 | 2022 | - | PROTEIN:potassium transport system protein kup |
| LCRIS_00162 | LCRIS_00162 | 167039 | 167473 | 435 | - | PROTEIN:hypothetical protein |
| LCRIS_00163 | LCRIS_00163 | 167805 | 168041 | 237 | + | PROTEIN:hypothetical protein |
| LCRIS_00164 | LCRIS_00164 | 168034 | 168369 | 336 | + | PROTEIN:hypothetical protein |
| LCRIS_00165 | LCRIS_00165 | 168436 | 168642 | 207 | + | PROTEIN:hypothetical protein |
| LCRIS_00166 | LCRIS_00166 | 168787 | 169125 | 339 | - | PROTEIN:transcriptional modulator of maze/toxin, m |
| LCRIS_00167 | LCRIS_00167 | 169148 | 169423 | 276 | - | PROTEIN:hypothetical protein |
| LCRIS_00168 | LCRIS_00168 | 169528 | 169947 | 420 | - | PROTEIN:hypothetical protein |
| LCRIS_00169 | LCRIS_00169 | 170098 | 170313 | 216 | + | PROTEIN:hypothetical protein |
| LCRIS_00170 | LCRIS_00170 | 170348 | 170620 | 273 | + | PROTEIN:hypothetical protein |
| LCRIS_00171 | slp1 | 171145 | 172545 | 1401 | + | PROTEIN:s-layer protein |
| LCRIS_00172 | LCRIS_00172 | 173088 | 174203 | 1116 | + | PROTEIN:transcriptional regulator |
| LCRIS_00173 | LCRIS_00173 | 174360 | 174779 | 420 | - | PROTEIN:hypothetical protein |
| LCRIS_00174 | LCRIS_00174 | 175321 | 175812 | 492 | - | PROTEIN:RNA polymerase sigma factor |
| LCRIS_00175 | LCRIS_00175 | 176025 | 176894 | 870 | - | PROTEIN:hypothetical protein |
| LCRIS_00176 | LCRIS_00176 | 177087 | 177980 | 894 | + | PROTEIN:hypothetical protein |
| LCRIS_00177 | slp2 | 178819 | 180291 | 1473 | - | PROTEIN:s-layer protein |
| LCRIS_00178 | LCRIS_00178 | 180435 | 180872 | 438 | + | PROTEIN:transposase DNA-binding domain family |
| LCRIS_00179 | LCRIS_00179 | 181240 | 181395 | 156 | + | PROTEIN:hypothetical protein |
| LCRIS_00180 | cadD | 181997 | 182617 | 621 | + | PROTEIN:cadmium resistance transporter |
| LCRIS_00181 | LCRIS_00181 | 182799 | 182990 | 192 | + | PROTEIN:hypothetical protein |
| LCRIS_00182 | LCRIS_00182 | 183149 | 183307 | 159 | - | PROTEIN:hypothetical protein |
| LCRIS_00183 | LCRIS_00183 | 183481 | 184707 | 1227 | + | PROTEIN:N-acetylmuramidase |
| LCRIS_00184 | cwlA | 184727 | 185815 | 1089 | + | PROTEIN:autolysin, amidase |
| LCRIS_00185 | LCRIS_00185 | 185904 | 187742 | 1839 | + | PROTEIN:Na+/H+ antiporter |
| LCRIS_00186 | pcp | 187787 | 188434 | 648 | - | PROTEIN:pyrrolidone-carboxylate peptidase |
| LCRIS_00187 | LCRIS_00187 | 188468 | 189433 | 966 | - | PROTEIN:oxidoreductase |
| LCRIS_00188 | LCRIS_00188 | 189441 | 190247 | 807 | - | PROTEIN:DNA polymerase |
| LCRIS_00189 | LCRIS_00189 | 190332 | 191087 | 756 | - | PROTEIN:hypothetical protein |
| LCRIS_00190 | gpmA1 | 191243 | 191935 | 693 | + | PROTEIN:2,3-bisphosphoglycerate-dependent phosphog |
| LCRIS_00191 | LCRIS_00191 | 192213 | 192773 | 561 | + | PROTEIN:aldose 1-epimerase |
| LCRIS_00192 | LCRIS_00192 | 192879 | 193979 | 1101 | + | PROTEIN:transcriptional regulator |
| LCRIS_00193 | LCRIS_00193 | 193979 | 194593 | 615 | + | PROTEIN:acyl-phosphate glycerol-3-phosphate acyltr |
| LCRIS_00194 | LCRIS_00194 | 194631 | 197621 | 2991 | + | PROTEIN:glucan modifying protein |
| LCRIS_00195 | LCRIS_00195 | 197723 | 199117 | 1395 | + | PROTEIN:fibronectin domain |
| LCRIS_00196 | tyrS | 199371 | 200633 | 1263 | + | PROTEIN:tyrosyl-tRNA synthetase |
| LCRIS_00197 | LCRIS_00197 | 201059 | 201415 | 357 | + | PROTEIN:hypothetical protein |
| LCRIS_02041 | tRNA-Thr | 201534 | 201606 | 73 | + | RNA:Thr tRNA |
| LCRIS_00198 | pepC1 | 201641 | 202954 | 1314 | + | PROTEIN:aminopeptidase c |
| LCRIS_00199 | LCRIS_00199 | 202954 | 203607 | 654 | + | PROTEIN:hd superfamily hydrolase |
| LCRIS_00200 | LCRIS_00200 | 203775 | 205394 | 1620 | + | PROTEIN:oligopeptide ABC transporter, oligopeptide |
| LCRIS_00201 | LCRIS_00201 | 205583 | 207211 | 1629 | + | PROTEIN:oligopeptide ABC transporter, oligopeptide |
| LCRIS_00202 | guaB | 207384 | 208526 | 1143 | + | PROTEIN:inosine-5'-monophosphate dehydrogenase |
| LCRIS_00203 | oppB1 | 208695 | 209624 | 930 | + | PROTEIN:oligopeptide ABC transporter, permease pro |
| LCRIS_00204 | oppC1 | 209633 | 210664 | 1032 | + | PROTEIN:oligopeptide ABC transporter, permease pro |
| LCRIS_00205 | oppD1 | 210680 | 211738 | 1059 | + | PROTEIN:oligopeptide ABC transporter, ATP-binding |
| LCRIS_00206 | oppF1 | 211759 | 212703 | 945 | + | PROTEIN:oligopeptide ABC transporter, ATP-binding |
| LCRIS_00207 | pepC2 | 212799 | 214115 | 1317 | - | PROTEIN:aminopeptidase c |
| LCRIS_00208 | hsp | 214257 | 214682 | 426 | - | PROTEIN:heat shock protein hsp20 |
| LCRIS_00209 | greA1 | 214924 | 215385 | 462 | + | PROTEIN:transcription elongation factor |
| LCRIS_00210 | LCRIS_00210 | 215465 | 216127 | 663 | + | PROTEIN:abortive infection protein |
| LCRIS_02098 | tRNA-Gly | 216175 | 216245 | 71 | - | RNA:Gly tRNA |
| LCRIS_00211 | LCRIS_00211 | 216255 | 216848 | 594 | - | PROTEIN:hypothetical protein |
| LCRIS_00212 | trpS | 216966 | 217988 | 1023 | + | PROTEIN:tryptophanyl-tRNA synthetase |
| LCRIS_00213 | comEB | 218004 | 218483 | 480 | + | PROTEIN:dcmp deaminase |
| LCRIS_00214 | LCRIS_00214 | 218497 | 219081 | 585 | + | PROTEIN:beta-propeller domain of methanol dehydrog |
| LCRIS_00215 | mgtA | 219380 | 222046 | 2667 | + | PROTEIN:magnesium-translocating p-type atpase |
| LCRIS_00216 | metG | 222145 | 224121 | 1977 | + | PROTEIN:methionyl-tRNA synthetase |
| LCRIS_00217 | tatD | 224121 | 224888 | 768 | + | PROTEIN:hydrolase, tatd family |
| LCRIS_00218 | rnmV | 224875 | 225441 | 567 | + | PROTEIN:primase-related protein |
| LCRIS_00219 | ksgA | 225431 | 226315 | 885 | + | PROTEIN:dimethyladenosine transferase |
| LCRIS_00220 | LCRIS_00220 | 226453 | 227859 | 1407 | + | PROTEIN:transposase |
| LCRIS_00221 | veg | 227983 | 228240 | 258 | + | PROTEIN:veg protein |
| LCRIS_00222 | purR | 228349 | 229179 | 831 | + | PROTEIN:pur operon repressor |
| LCRIS_00223 | glmU | 229226 | 230611 | 1386 | + | PROTEIN:bifunctional protein glmu |
| LCRIS_00224 | LCRIS_00224 | 230740 | 231492 | 753 | + | PROTEIN:hypothetical protein |
| LCRIS_00225 | LCRIS_00225 | 231623 | 233392 | 1770 | + | PROTEIN:cell separation protein |
| LCRIS_00226 | prs2 | 233620 | 234594 | 975 | + | PROTEIN:ribose-phosphate pyrophosphokinase |
| LCRIS_00227 | LCRIS_00227 | 234762 | 235682 | 921 | + | PROTEIN:transcriptional regulator |
| LCRIS_00228 | LCRIS_00228 | 235743 | 236021 | 279 | - | PROTEIN:hypothetical protein |
| LCRIS_00229 | LCRIS_00229 | 236053 | 237426 | 1374 | - | PROTEIN:hd superfamily phosphohydrolase |
| LCRIS_00230 | LCRIS_00230 | 237541 | 237945 | 405 | + | PROTEIN:hypothetical protein |
| LCRIS_00231 | rpoE | 237992 | 238549 | 558 | + | PROTEIN:DNA-directed RNA polymerase, delta subunit |
| LCRIS_00232 | pyrG | 238666 | 240285 | 1620 | + | PROTEIN:ctp synthase |
| LCRIS_00233 | murA | 240420 | 241715 | 1296 | + | PROTEIN:UDP-N-acetylglucosamine 1-carboxyvinyltran |
| LCRIS_00234 | LCRIS_00234 | 241784 | 242305 | 522 | + | PROTEIN:acetyltransferase, gnat family |
| LCRIS_00235 | pepDA | 242361 | 243785 | 1425 | - | PROTEIN:dipeptidase |
| LCRIS_00236 | LCRIS_00236 | 243872 | 244684 | 813 | - | PROTEIN:raffinose operon transcriptional regulator |
| LCRIS_00237 | pbuX | 244831 | 246117 | 1287 | - | PROTEIN:xanthine permease |
| LCRIS_00238 | xpt | 246124 | 246702 | 579 | - | PROTEIN:xanthine phosphoribosyltransferase |
| LCRIS_00239 | LCRIS_00239 | 246936 | 247109 | 174 | - | PROTEIN:hypothetical protein |
| LCRIS_00240 | LCRIS_00240 | 247131 | 247313 | 183 | - | PROTEIN:transcriptional regulator |
| LCRIS_00241 | guaA | 247536 | 249086 | 1551 | + | PROTEIN:gmp synthase (glutamine-hydrolyzing) |
| LCRIS_00242 | LCRIS_00242 | 249584 | 249775 | 192 | + | PROTEIN:hypothetical protein |
| LCRIS_00243 | LCRIS_00243 | 249941 | 250495 | 555 | + | PROTEIN:transposase |
| LCRIS_00244 | LCRIS_00244 | 250783 | 256479 | 5697 | + | PROTEIN:protein with ysrik-signal peptide |
| LCRIS_00245 | LCRIS_00245 | 256936 | 257262 | 327 | - | PROTEIN:transcriptional regulator |
| LCRIS_00246 | LCRIS_00246 | 257262 | 257564 | 303 | + | PROTEIN:hypothetical protein |
| LCRIS_00247 | LCRIS_00247 | 257688 | 257876 | 189 | - | PROTEIN:hypothetical protein |
| LCRIS_00248 | LCRIS_00248 | 258004 | 258339 | 336 | - | PROTEIN:transcriptional regulator |
| LCRIS_00249 | LCRIS_00249 | 258489 | 259511 | 1023 | + | PROTEIN:alcohol dehydrogenase, zinc-binding |
| LCRIS_00250 | LCRIS_00250 | 259508 | 261277 | 1770 | - | PROTEIN:transposase |
| LCRIS_00251 | LCRIS_00251 | 261774 | 262346 | 573 | - | PROTEIN:transposase |
| LCRIS_00252 | LCRIS_00252 | 262343 | 263065 | 723 | - | PROTEIN:transposase, is605-tnpb |
| LCRIS_00253 | LCRIS_00253 | 263096 | 263710 | 615 | - | PROTEIN:resolvase, n terminal domain family protei |
| LCRIS_00254 | ogt2 | 263980 | 264489 | 510 | - | PROTEIN:methylated-DNA--protein-cysteine methyltra |
| LCRIS_00255 | LCRIS_00255 | 264559 | 265074 | 516 | - | PROTEIN:hypothetical protein |
| LCRIS_00256 | LCRIS_00256 | 265220 | 265597 | 378 | + | PROTEIN:hypothetical protein |
| LCRIS_00257 | LCRIS_00257 | 265624 | 266184 | 561 | + | PROTEIN:resolvase, n terminal domain family protei |
| LCRIS_00258 | LCRIS_00258 | 266215 | 267507 | 1293 | + | PROTEIN:transposase |
| LCRIS_00259 | LCRIS_00259 | 267539 | 267823 | 285 | + | PROTEIN:hypothetical protein |
| LCRIS_00260 | LCRIS_00260 | 267795 | 268631 | 837 | + | PROTEIN:chloride channel protein |
| LCRIS_00261 | LCRIS_00261 | 268644 | 269645 | 1002 | + | PROTEIN:glycolate oxidase |
| LCRIS_00262 | LCRIS_00262 | 269967 | 270302 | 336 | + | PROTEIN:hypothetical protein |
| LCRIS_00263 | LCRIS_00263 | 270283 | 270558 | 276 | + | PROTEIN:hypothetical protein |
| LCRIS_00264 | glyA | 270617 | 271852 | 1236 | + | PROTEIN:serine hydroxymethyltransferase |
| LCRIS_00265 | LCRIS_00265 | 271956 | 273548 | 1593 | + | PROTEIN:amino acid permease |
| LCRIS_00266 | LCRIS_00266 | 273594 | 274421 | 828 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_00267 | dexB | 274537 | 276156 | 1620 | + | PROTEIN:alpha,alpha-phosphotrehalase |
| LCRIS_00268 | rpmE | 276266 | 276511 | 246 | + | PROTEIN:50S ribosomal protein l31 type b |
| LCRIS_00269 | murF | 276637 | 278004 | 1368 | + | PROTEIN:UDP-N-acetylmuramoyl-tripeptide--D-alanyl- |
| LCRIS_00270 | deaD | 278018 | 279502 | 1485 | + | PROTEIN:ATP-dependent RNA helicase |
| LCRIS_00271 | acpS | 279585 | 279941 | 357 | + | PROTEIN:holo-[acyl-carrier-protein] synthase |
| LCRIS_00272 | alr | 279946 | 281076 | 1131 | + | PROTEIN:alanine racemase |
| LCRIS_00273 | LCRIS_00273 | 281141 | 281848 | 708 | - | PROTEIN:cbs domain protein |
| LCRIS_00274 | ldh1 | 282019 | 282990 | 972 | - | PROTEIN:L-lactate dehydrogenase |
| LCRIS_00275 | pth | 283127 | 283684 | 558 | + | PROTEIN:peptidyl-tRNA hydrolase |
| LCRIS_00276 | mfd | 283686 | 287180 | 3495 | + | PROTEIN:transcription-repair coupling factor |
| LCRIS_00277 | LCRIS_00277 | 287196 | 287468 | 273 | + | PROTEIN:RNA-binding s4 domain protein |
| LCRIS_00278 | LCRIS_00278 | 287566 | 288972 | 1407 | + | PROTEIN:transposase |
| LCRIS_00279 | LCRIS_00279 | 289112 | 289489 | 378 | + | PROTEIN:septum formation initiator |
| LCRIS_00280 | LCRIS_00280 | 289489 | 289851 | 363 | + | PROTEIN:ribosomal protein s1 |
| LCRIS_00281 | mesJ | 289890 | 291143 | 1254 | + | PROTEIN:cell cycle protein |
| LCRIS_00282 | ftsH | 291238 | 293406 | 2169 | + | PROTEIN:cell division protein ftsh |
| LCRIS_00283 | hslO | 293459 | 294349 | 891 | + | PROTEIN:33 kda chaperonin |
| LCRIS_00284 | dusB | 294427 | 295452 | 1026 | + | PROTEIN:tRNA-dihydrouridine synthase |
| LCRIS_00285 | lysS | 295468 | 297015 | 1548 | + | PROTEIN:lysyl-tRNA synthetase |
| LCRIS_00286 | LCRIS_00286 | 297354 | 297914 | 561 | + | PROTEIN:hypothetical protein |
| LCRIS_00287 | LCRIS_00287 | 298495 | 298965 | 471 | + | PROTEIN:hypothetical protein |
| LCRIS_02042 | tRNA-Gly | 299102 | 299176 | 75 | + | RNA:Gly tRNA |
| LCRIS_00288 | LCRIS_00288 | 299388 | 301247 | 1860 | + | PROTEIN:5'-nucleotidase |
| LCRIS_02043 | tRNA-Gly | 301350 | 301424 | 75 | + | RNA:Gly tRNA |
| LCRIS_02044 | tRNA-Gly | 301536 | 301610 | 75 | + | RNA:Gly tRNA |
| LCRIS_02045 | tRNA-Pro | 301648 | 301721 | 74 | + | RNA:Pro tRNA |
| LCRIS_00289 | LCRIS_00289 | 301796 | 302251 | 456 | + | PROTEIN:gaf domain protein |
| LCRIS_00290 | clpC | 302356 | 304836 | 2481 | + | PROTEIN:atpase aaa-2 domain protein |
| LCRIS_00291 | rpoB | 305058 | 308696 | 3639 | + | PROTEIN:DNA-directed RNA polymerase, beta subunit |
| LCRIS_00292 | rpoC | 308712 | 312371 | 3660 | + | PROTEIN:DNA-directed RNA polymerase, beta' subunit |
| LCRIS_00293 | comC | 312434 | 313117 | 684 | - | PROTEIN:type iv prepilin peptidase |
| LCRIS_00294 | rpsL | 313299 | 313706 | 408 | + | PROTEIN:30S ribosomal protein s12 |
| LCRIS_00295 | rpsG | 313722 | 314192 | 471 | + | PROTEIN:30S ribosomal protein s7 |
| LCRIS_00296 | fusA | 314226 | 316319 | 2094 | + | PROTEIN:elongation factor g |
| LCRIS_00297 | rpsJ | 316635 | 316943 | 309 | + | PROTEIN:30S ribosomal protein s10 |
| LCRIS_00298 | rplC | 316967 | 317605 | 639 | + | PROTEIN:50S ribosomal protein l3 |
| LCRIS_00299 | rplD | 317620 | 318237 | 618 | + | PROTEIN:50S ribosomal protein l4 |
| LCRIS_00300 | rplW | 318237 | 318542 | 306 | + | PROTEIN:50S ribosomal protein l23 |
| LCRIS_00301 | rplB | 318558 | 319394 | 837 | + | PROTEIN:50S ribosomal protein l2 |
| LCRIS_00302 | rpsS | 319413 | 319697 | 285 | + | PROTEIN:30S ribosomal protein s19 |
| LCRIS_00303 | rplV | 319717 | 320070 | 354 | + | PROTEIN:50S ribosomal protein l22 |
| LCRIS_00304 | rpsC | 320084 | 320758 | 675 | + | PROTEIN:30S ribosomal protein s3 |
| LCRIS_00305 | rplP | 320758 | 321198 | 441 | + | PROTEIN:50S ribosomal protein l16 |
| LCRIS_00306 | rpmC | 321188 | 321385 | 198 | + | PROTEIN:50S ribosomal protein l29 |
| LCRIS_00307 | rpsQ | 321404 | 321670 | 267 | + | PROTEIN:30S ribosomal protein s17 |
| LCRIS_00308 | rplN | 321702 | 322070 | 369 | + | PROTEIN:50S ribosomal protein l14 |
| LCRIS_00309 | rplX | 322090 | 322326 | 237 | + | PROTEIN:50S ribosomal protein l24 |
| LCRIS_00310 | rplE | 322341 | 322883 | 543 | + | PROTEIN:50S ribosomal protein l5 |
| LCRIS_00311 | rpsZ | 322896 | 323081 | 186 | + | PROTEIN:30S ribosomal protein s14 type z |
| LCRIS_00312 | rpsH | 323106 | 323504 | 399 | + | PROTEIN:30S ribosomal protein s8 |
| LCRIS_00313 | rplF | 323529 | 324059 | 531 | + | PROTEIN:50S ribosomal protein l6 |
| LCRIS_00314 | rplR | 324086 | 324442 | 357 | + | PROTEIN:50S ribosomal protein l18 |
| LCRIS_00315 | rpsE | 324460 | 324966 | 507 | + | PROTEIN:30S ribosomal protein s5 |
| LCRIS_00316 | rpmD | 324984 | 325169 | 186 | + | PROTEIN:50S ribosomal protein l30 |
| LCRIS_00317 | rplO | 325193 | 325633 | 441 | + | PROTEIN:50S ribosomal protein l15 |
| LCRIS_00318 | secY | 325633 | 326928 | 1296 | + | PROTEIN:preprotein translocase secy subunit |
| LCRIS_00319 | adk | 326939 | 327595 | 657 | + | PROTEIN:adenylate kinase |
| LCRIS_00320 | infA | 327671 | 327892 | 222 | + | PROTEIN:translation initiation factor if-1 |
| LCRIS_00321 | rpsM | 328046 | 328396 | 351 | + | PROTEIN:30S ribosomal protein s13 |
| LCRIS_00322 | rpsK | 328421 | 328810 | 390 | + | PROTEIN:30S ribosomal protein s11 |
| LCRIS_00323 | rpoA | 328856 | 329794 | 939 | + | PROTEIN:DNA-directed RNA polymerase, alpha subunit |
| LCRIS_00324 | rplQ | 329822 | 330205 | 384 | + | PROTEIN:50S ribosomal protein l17 |
| LCRIS_00325 | cbiO2 | 330389 | 331240 | 852 | + | PROTEIN:cobalt import ATP-binding protein cbio 1 |
| LCRIS_00326 | cbiO3 | 331216 | 332061 | 846 | + | PROTEIN:cobalt import ATP-binding protein cbio 2 |
| LCRIS_00327 | cbiQ2 | 332065 | 332862 | 798 | + | PROTEIN:cobalt ABC transporter permease |
| LCRIS_00328 | truA | 332867 | 333655 | 789 | + | PROTEIN:tRNA pseudouridine synthase a |
| LCRIS_00329 | rplM | 333760 | 334203 | 444 | + | PROTEIN:50S ribosomal protein l13 |
| LCRIS_00330 | rpsI | 334215 | 334610 | 396 | + | PROTEIN:30S ribosomal protein s9 |
| LCRIS_00331 | LCRIS_00331 | 334862 | 335170 | 309 | + | PROTEIN:hypothetical protein |
| LCRIS_00332 | LCRIS_00332 | 335183 | 336340 | 1158 | + | PROTEIN:hypothetical protein |
| LCRIS_00333 | LCRIS_00333 | 336562 | 337011 | 450 | + | PROTEIN:acetyltransferase, gnat family |
| LCRIS_00334 | thiJ | 337040 | 337624 | 585 | - | PROTEIN:dj-1 family protease |
| LCRIS_00335 | LCRIS_00335 | 337661 | 338320 | 660 | - | PROTEIN:hypothetical protein |
| LCRIS_00336 | LCRIS_00336 | 338394 | 339152 | 759 | + | PROTEIN:predicted permease |
| LCRIS_00337 | gpm1 | 339196 | 339876 | 681 | + | PROTEIN:phosphoglycerate mutase |
| LCRIS_00338 | LCRIS_00338 | 339937 | 340836 | 900 | + | PROTEIN:cation efflux protein |
| LCRIS_00339 | LCRIS_00339 | 340845 | 341027 | 183 | + | PROTEIN:hypothetical protein |
| LCRIS_00340 | gpmA2 | 341039 | 341719 | 681 | + | PROTEIN:2,3-bisphosphoglycerate-dependent phosphog |
| LCRIS_00341 | LCRIS_00341 | 341830 | 342765 | 936 | + | PROTEIN:kinase |
| LCRIS_00342 | LCRIS_00342 | 342829 | 343077 | 249 | - | PROTEIN:hypothetical protein |
| LCRIS_00343 | LCRIS_00343 | 343074 | 343235 | 162 | - | PROTEIN:hypothetical protein |
| LCRIS_00344 | pepC3 | 343375 | 344724 | 1350 | - | PROTEIN:aminopeptidase c |
| LCRIS_00345 | LCRIS_00345 | 344792 | 345091 | 300 | - | PROTEIN:hypothetical protein |
| LCRIS_00346 | dut | 345223 | 345774 | 552 | + | PROTEIN:deoxyuridine 5'-triphosphate nucleotidohyd |
| LCRIS_00347 | radA | 345774 | 347150 | 1377 | + | PROTEIN:DNA repair protein rada |
| LCRIS_00348 | gltX | 347228 | 348727 | 1500 | + | PROTEIN:glutamyl-tRNA synthetase |
| LCRIS_00349 | cysS | 348818 | 350251 | 1434 | + | PROTEIN:cysteinyl-tRNA synthetase |
| LCRIS_00350 | LCRIS_00350 | 350244 | 350687 | 444 | + | PROTEIN:ribonuclease iii |
| LCRIS_00351 | yjfH | 350674 | 351429 | 756 | + | PROTEIN:RNA methyltransferase, trmh family, group |
| LCRIS_00352 | LCRIS_00352 | 351563 | 352108 | 546 | + | PROTEIN:DNA-directed RNA polymerase sigma factor, |
| LCRIS_00353 | LCRIS_00353 | 352172 | 352390 | 219 | + | PROTEIN:hypothetical protein |
| LCRIS_00354 | gntK | 352507 | 354015 | 1509 | + | PROTEIN:gluconate kinase |
| LCRIS_00355 | dxs | 354062 | 355807 | 1746 | - | PROTEIN:1-deoxy-d-xylulose-5-phosphate synthase |
| LCRIS_00356 | rpmG1 | 355993 | 356142 | 150 | + | PROTEIN:50S ribosomal protein l33 |
| LCRIS_00357 | secE | 356152 | 356322 | 171 | + | PROTEIN:preprotein translocase, sece subunit |
| LCRIS_00358 | nusG | 356431 | 356988 | 558 | + | PROTEIN:transcription antitermination protein nusg |
| LCRIS_00359 | rplK | 357119 | 357544 | 426 | + | PROTEIN:50S ribosomal protein l11 |
| LCRIS_00360 | rplA | 357621 | 358313 | 693 | + | PROTEIN:50S ribosomal protein l1 |
| LCRIS_00361 | pstS | 358433 | 359299 | 867 | + | PROTEIN:phosphate ABC transporter, phosphate-bindi |
| LCRIS_00362 | pstC | 359303 | 360298 | 996 | + | PROTEIN:phosphate ABC transporter, permease protei |
| LCRIS_00363 | pstA | 360300 | 361187 | 888 | + | PROTEIN:phosphate ABC transporter, permease protei |
| LCRIS_00364 | pstB1 | 361196 | 361990 | 795 | + | PROTEIN:phosphate ABC transporter, ATP-binding pro |
| LCRIS_00365 | pstB2 | 361992 | 362747 | 756 | + | PROTEIN:phosphate ABC transporter, ATP-binding pro |
| LCRIS_00366 | phoU | 362765 | 363442 | 678 | + | PROTEIN:phosphate transport system regulatory prot |
| LCRIS_00367 | LCRIS_00367 | 363503 | 363658 | 156 | - | PROTEIN:hypothetical protein |
| LCRIS_00368 | LCRIS_00368 | 363648 | 364136 | 489 | - | PROTEIN:transposase |
| LCRIS_00369 | LCRIS_00369 | 364218 | 364496 | 279 | + | PROTEIN:hypothetical protein |
| LCRIS_00370 | rplJ | 364700 | 365212 | 513 | + | PROTEIN:50S ribosomal protein l10 |
| LCRIS_00371 | rplL | 365265 | 365627 | 363 | + | PROTEIN:50S ribosomal protein l7/l12 |
| LCRIS_00372 | LCRIS_00372 | 365704 | 366546 | 843 | + | PROTEIN:k channel |
| LCRIS_00373 | LCRIS_00373 | 366552 | 367172 | 621 | + | PROTEIN:methyltransferase small |
| LCRIS_00374 | LCRIS_00374 | 367300 | 367482 | 183 | + | PROTEIN:hypothetical protein |
| LCRIS_00375 | LCRIS_00375 | 367557 | 368057 | 501 | + | PROTEIN:tRNA-specific adenosine deaminase |
| LCRIS_00376 | dnaX | 368263 | 370068 | 1806 | + | PROTEIN:DNA polymerase iii, gamma/tau subunit |
| LCRIS_00377 | LCRIS_00377 | 370075 | 370665 | 591 | + | PROTEIN:hypothetical protein |
| LCRIS_00378 | LCRIS_00378 | 370674 | 370904 | 231 | + | PROTEIN:hypothetical protein |
| LCRIS_00379 | LCRIS_00379 | 370920 | 371255 | 336 | + | PROTEIN:hypothetical protein |
| LCRIS_00380 | recR | 371256 | 371855 | 600 | + | PROTEIN:recombination protein recr |
| LCRIS_00381 | LCRIS_00381 | 371855 | 372094 | 240 | + | PROTEIN:hypothetical protein |
| LCRIS_00382 | tmk | 372197 | 372835 | 639 | + | PROTEIN:thymidylate kinase |
| LCRIS_00383 | LCRIS_00383 | 372848 | 373168 | 321 | + | PROTEIN:hypothetical protein |
| LCRIS_00384 | holB | 373168 | 374025 | 858 | + | PROTEIN:DNA polymerase iii, delta subunit |
| LCRIS_00385 | LCRIS_00385 | 374035 | 374385 | 351 | + | PROTEIN:initiation-control protein yaba |
| LCRIS_00386 | LCRIS_00386 | 374385 | 375236 | 852 | + | PROTEIN:tetrapyrrole methylase family protein |
| LCRIS_00387 | LCRIS_00387 | 375239 | 375973 | 735 | + | PROTEIN:acyl-acp thioesterase |
| LCRIS_00388 | LCRIS_00388 | 376029 | 376547 | 519 | + | PROTEIN:hypothetical protein |
| LCRIS_00389 | LCRIS_00389 | 376577 | 377311 | 735 | + | PROTEIN:glycoprotein endopeptidase |
| LCRIS_00390 | rimI | 377295 | 377867 | 573 | + | PROTEIN:ribosomal-protein-alanine acetyltransferas |
| LCRIS_00391 | gcp | 377864 | 378913 | 1050 | + | PROTEIN:o-sialoglycoprotein endopeptidase |
| LCRIS_00392 | LCRIS_00392 | 379153 | 380379 | 1227 | - | PROTEIN:transposase |
| LCRIS_00393 | LCRIS_00393 | 380645 | 380926 | 282 | + | PROTEIN:transposase |
| LCRIS_00394 | LCRIS_00394 | 380932 | 381168 | 237 | + | PROTEIN:transposase (is150) |
| LCRIS_00395 | LCRIS_00395 | 381192 | 381809 | 618 | + | PROTEIN:transposase |
| LCRIS_00396 | LCRIS_00396 | 381857 | 383779 | 1923 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00397 | rex | 383954 | 384589 | 636 | + | PROTEIN:redox-sensing transcriptional repressor re |
| LCRIS_00398 | scrR | 384627 | 385613 | 987 | - | PROTEIN:sucrose operon repressor scrr |
| LCRIS_00399 | scrB | 385629 | 387086 | 1458 | - | PROTEIN:sucrose-6-phosphate hydrolase |
| LCRIS_00400 | scrA1 | 387296 | 389245 | 1950 | + | PROTEIN:PTS system sucrose-specific iiABC componen |
| LCRIS_00401 | LCRIS_00401 | 389401 | 390369 | 969 | + | PROTEIN:hypothetical protein |
| LCRIS_00402 | LCRIS_00402 | 390415 | 390930 | 516 | + | PROTEIN:hypothetical protein |
| LCRIS_00403 | groES | 391112 | 391396 | 285 | + | PROTEIN:10 kda chaperonin |
| LCRIS_00404 | groEL | 391450 | 393075 | 1626 | + | PROTEIN:60 kda chaperonin |
| LCRIS_00405 | mutS1 | 393334 | 395910 | 2577 | + | PROTEIN:DNA mismatch repair protein muts |
| LCRIS_00406 | mutL | 395910 | 397835 | 1926 | + | PROTEIN:DNA mismatch repair protein mutl |
| LCRIS_00407 | ruvA | 397835 | 398422 | 588 | + | PROTEIN:holliday junction ATP-dependent DNA helica |
| LCRIS_00408 | ruvB | 398462 | 399481 | 1020 | + | PROTEIN:holliday junction ATP-dependent DNA helica |
| LCRIS_00409 | yajC | 399541 | 399933 | 393 | + | PROTEIN:protein translocase subunit yajc |
| LCRIS_00410 | zwf | 400037 | 401488 | 1452 | + | PROTEIN:glucose-6-phosphate 1-dehydrogenase |
| LCRIS_00411 | dinB | 401364 | 402599 | 1236 | + | PROTEIN:DNA polymerase iv |
| LCRIS_00412 | LCRIS_00412 | 402663 | 403619 | 957 | + | PROTEIN:phosphoesterase, dhh family protein |
| LCRIS_00413 | LCRIS_00413 | 403612 | 404973 | 1362 | + | PROTEIN:ATP-dependent RNA helicase |
| LCRIS_00414 | alaS | 405216 | 407855 | 2640 | + | PROTEIN:alanyl-tRNA synthetase |
| LCRIS_00415 | LCRIS_00415 | 407916 | 408173 | 258 | + | PROTEIN:hypothetical protein |
| LCRIS_00416 | ruvX | 408173 | 408601 | 429 | + | PROTEIN:holliday junction resolvase |
| LCRIS_00417 | LCRIS_00417 | 408603 | 408914 | 312 | + | PROTEIN:hypothetical protein |
| LCRIS_00418 | mutS2 | 408972 | 411329 | 2358 | + | PROTEIN:DNA mismatch repair protein muts2 |
| LCRIS_00419 | trxA | 411412 | 411723 | 312 | + | PROTEIN:thioredoxin |
| LCRIS_00420 | mscL | 411777 | 412151 | 375 | - | PROTEIN:large-conductance mechanosensitive channel |
| LCRIS_00421 | LCRIS_00421 | 412240 | 412662 | 423 | - | PROTEIN:hydrocarbon binding protein, v4r domain |
| LCRIS_00422 | murI | 412739 | 413545 | 807 | + | PROTEIN:glutamate racemase |
| LCRIS_00423 | rdgB | 413545 | 414165 | 621 | + | PROTEIN:nucleoside-triphosphatase |
| LCRIS_00424 | LCRIS_00424 | 414260 | 415228 | 969 | + | PROTEIN:hypothetical protein |
| LCRIS_00425 | LCRIS_00425 | 415379 | 416035 | 657 | + | PROTEIN:hypothetical protein |
| LCRIS_00426 | LCRIS_00426 | 416438 | 417274 | 837 | - | PROTEIN:mechanosensitive ion channel |
| LCRIS_00427 | LCRIS_00427 | 417383 | 417808 | 426 | + | PROTEIN:methyl-accepting chemotaxis-like protein |
| LCRIS_00428 | LCRIS_00428 | 417826 | 418194 | 369 | + | PROTEIN:hypothetical protein |
| LCRIS_00429 | pepQ | 418254 | 419360 | 1107 | - | PROTEIN:xaa-pro dipeptidase |
| LCRIS_00430 | ccpA | 419511 | 420512 | 1002 | + | PROTEIN:catabolite control protein a |
| LCRIS_00431 | LCRIS_00431 | 420554 | 421924 | 1371 | - | PROTEIN:glycerophosphoryl diester phosphodiesteras |
| LCRIS_00432 | LCRIS_00432 | 422038 | 422358 | 321 | + | PROTEIN:hypothetical protein |
| LCRIS_00433 | LCRIS_00433 | 422374 | 423756 | 1383 | + | PROTEIN:5'-nucleotidase/2',3'-cyclic phosphodieste |
| LCRIS_00434 | LCRIS_00434 | 423749 | 424339 | 591 | + | PROTEIN:hypothetical protein |
| LCRIS_00435 | LCRIS_00435 | 424339 | 425109 | 771 | + | PROTEIN:had superfamily hydrolase, subfamily iia |
| LCRIS_00436 | LCRIS_00436 | 425106 | 425714 | 609 | + | PROTEIN:hypothetical protein |
| LCRIS_00437 | dedA | 425726 | 426376 | 651 | - | PROTEIN:deda protein |
| LCRIS_00438 | trxB1 | 426386 | 427378 | 993 | - | PROTEIN:thioredoxin reductase |
| LCRIS_02036 | 16srRNA | 427906 | 429457 | 1552 | + | RNA:16S ribosomal RNA |
| LCRIS_00439 | LCRIS_00439 | 427983 | 428108 | 126 | - | PROTEIN:hypothetical protein |
| LCRIS_00440 | LCRIS_00440 | 428168 | 428602 | 435 | - | PROTEIN:pg1 protein, homology to Homo sapiens |
| LCRIS_00441 | LCRIS_00441 | 429036 | 429389 | 354 | + | PROTEIN:hypothetical protein |
| LCRIS_02046 | tRNA-Ile | 429594 | 429668 | 75 | + | RNA:Ile tRNA |
| LCRIS_02047 | tRNA-Ala | 429725 | 429797 | 73 | + | RNA:Ala tRNA |
| LCRIS_02032 | 23srRNA | 429920 | 432825 | 2906 | + | RNA:23S ribosomal RNA |
| LCRIS_00442 | LCRIS_00442 | 430474 | 430944 | 471 | - | PROTEIN:hypothetical protein |
| LCRIS_00443 | LCRIS_00443 | 432271 | 432558 | 288 | + | PROTEIN:hypothetical protein |
| LCRIS_02028 | 5srRNA | 432898 | 433013 | 116 | + | RNA:5S ribosomal RNA |
| LCRIS_02048 | tRNA-Val | 433020 | 433092 | 73 | + | RNA:Val tRNA |
| LCRIS_02049 | tRNA-Lys | 433097 | 433169 | 73 | + | RNA:Lys tRNA |
| LCRIS_02050 | tRNA-Leu | 433183 | 433264 | 82 | + | RNA:Leu tRNA |
| LCRIS_02051 | tRNA-Thr | 433289 | 433361 | 73 | + | RNA:Thr tRNA |
| LCRIS_02052 | tRNA-Gly | 433372 | 433443 | 72 | + | RNA:Gly tRNA |
| LCRIS_02053 | tRNA-Leu | 433455 | 433540 | 86 | + | RNA:Leu tRNA |
| LCRIS_02054 | tRNA-Arg | 433549 | 433622 | 74 | + | RNA:Arg tRNA |
| LCRIS_02055 | tRNA-Pro | 433628 | 433701 | 74 | + | RNA:Pro tRNA |
| LCRIS_02056 | tRNA-Met | 433732 | 433805 | 74 | + | RNA:Met tRNA |
| LCRIS_02057 | tRNA-Met | 433819 | 433892 | 74 | + | RNA:Met tRNA |
| LCRIS_02058 | tRNA-Met | 433916 | 433989 | 74 | + | RNA:Met tRNA |
| LCRIS_02059 | tRNA-Asp | 433993 | 434066 | 74 | + | RNA:Asp tRNA |
| LCRIS_02060 | tRNA-Phe | 434073 | 434145 | 73 | + | RNA:Phe tRNA |
| LCRIS_02061 | tRNA-Gly | 434164 | 434234 | 71 | + | RNA:Gly tRNA |
| LCRIS_02062 | tRNA-Ile | 434241 | 434315 | 75 | + | RNA:Ile tRNA |
| LCRIS_02063 | tRNA-Ser | 434318 | 434410 | 93 | + | RNA:Ser tRNA |
| LCRIS_02064 | tRNA-Glu | 435164 | 435235 | 72 | + | RNA:Glu tRNA |
| LCRIS_02065 | tRNA-Ser | 435270 | 435356 | 87 | + | RNA:Ser tRNA |
| LCRIS_02066 | tRNA-Met | 435368 | 435441 | 74 | + | RNA:Met tRNA |
| LCRIS_02067 | tRNA-Asp | 435445 | 435518 | 74 | + | RNA:Asp tRNA |
| LCRIS_02068 | tRNA-Phe | 435525 | 435600 | 76 | + | RNA:Phe tRNA |
| LCRIS_02069 | tRNA-Tyr | 435604 | 435685 | 82 | + | RNA:Tyr tRNA |
| LCRIS_02070 | tRNA-Trp | 435690 | 435760 | 71 | + | RNA:Trp tRNA |
| LCRIS_02071 | tRNA-His | 435773 | 435848 | 76 | + | RNA:His tRNA |
| LCRIS_02072 | tRNA-Gln | 435853 | 435924 | 72 | + | RNA:Gln tRNA |
| LCRIS_02073 | tRNA-Cys | 435948 | 436018 | 71 | + | RNA:Cys tRNA |
| LCRIS_02074 | tRNA-Leu | 436060 | 436144 | 85 | + | RNA:Leu tRNA |
| LCRIS_00444 | LCRIS_00444 | 436264 | 437442 | 1179 | + | PROTEIN:glycosyl transferase, group 1 |
| LCRIS_00445 | LCRIS_00445 | 437443 | 438486 | 1044 | + | PROTEIN:glycosyltransferase |
| LCRIS_00446 | LCRIS_00446 | 438486 | 439511 | 1026 | + | PROTEIN:integral hypothetical protein |
| LCRIS_00447 | LCRIS_00447 | 439586 | 441646 | 2061 | + | PROTEIN:sulfatase |
| LCRIS_00448 | secG | 441714 | 441947 | 234 | + | PROTEIN:preprotein translocase |
| LCRIS_00449 | rnr | 442020 | 444359 | 2340 | + | PROTEIN:ribonuclease r |
| LCRIS_00450 | smpB | 444372 | 444830 | 459 | + | PROTEIN:ssra-binding protein |
| LCRIS_02035 | 16srRNA | 445456 | 447007 | 1552 | + | RNA:16S ribosomal RNA |
| LCRIS_00451 | LCRIS_00451 | 445668 | 445784 | 117 | - | PROTEIN:hypothetical protein |
| LCRIS_00452 | LCRIS_00452 | 445718 | 446152 | 435 | - | PROTEIN:pg1 protein, homology to Homo sapiens |
| LCRIS_00453 | LCRIS_00453 | 446333 | 446440 | 108 | - | PROTEIN:hypothetical protein |
| LCRIS_00454 | LCRIS_00454 | 446586 | 446939 | 354 | + | PROTEIN:hypothetical protein |
| LCRIS_02031 | 23srRNA | 447220 | 450125 | 2906 | + | RNA:23S ribosomal RNA |
| LCRIS_00455 | LCRIS_00455 | 447774 | 448244 | 471 | - | PROTEIN:hypothetical protein |
| LCRIS_00456 | LCRIS_00456 | 449571 | 449858 | 288 | + | PROTEIN:hypothetical protein |
| LCRIS_02027 | 5srRNA | 450198 | 450313 | 116 | + | RNA:5S ribosomal RNA |
| LCRIS_02075 | tRNA-Asn | 450326 | 450398 | 73 | + | RNA:Asn tRNA |
| LCRIS_00457 | manL | 450694 | 451701 | 1008 | + | PROTEIN:PTS system mannose-specific iiab component |
| LCRIS_00458 | manY1 | 451723 | 452532 | 810 | + | PROTEIN:PTS system mannose-specific iic component |
| LCRIS_00459 | manZ1 | 452561 | 453481 | 921 | + | PROTEIN:PTS system mannose-specific iid component |
| LCRIS_00460 | manO | 453485 | 453853 | 369 | + | PROTEIN:mano |
| LCRIS_00461 | LCRIS_00461 | 453896 | 454777 | 882 | - | PROTEIN:transcriptional regulator |
| LCRIS_00462 | maa1 | 454886 | 455416 | 531 | + | PROTEIN:maltose o-acetyltransferase |
| LCRIS_00463 | LCRIS_00463 | 455435 | 456808 | 1374 | - | PROTEIN:d-serine/D-alanine/glycine transporter |
| LCRIS_00464 | adhE | 457121 | 459745 | 2625 | + | PROTEIN:aldehyde-alcohol dehydrogenase |
| LCRIS_00465 | glmS | 459942 | 461753 | 1812 | + | PROTEIN:glucosamine--fructose-6-phosphate aminotra |
| LCRIS_00466 | mtlR | 461853 | 463907 | 2055 | + | PROTEIN:transcription regulator |
| LCRIS_00467 | mtlA | 464042 | 465823 | 1782 | + | PROTEIN:PTS system mannitol-specific iibc componen |
| LCRIS_00468 | mtlF | 465839 | 466291 | 453 | + | PROTEIN:PTS system mannitol-specific iia component |
| LCRIS_00469 | mtlD | 466294 | 467463 | 1170 | + | PROTEIN:mannitol-1-phosphate 5-dehydrogenase |
| LCRIS_00470 | LCRIS_00470 | 467624 | 468355 | 732 | + | PROTEIN:carboxymuconolactone decarboxylase |
| LCRIS_00471 | LCRIS_00471 | 468819 | 469232 | 414 | + | PROTEIN:nudix family hydrolase |
| LCRIS_00472 | LCRIS_00472 | 469378 | 470232 | 855 | - | PROTEIN:ranscriptional activator rgg/gadr/mutr-fam |
| LCRIS_00473 | LCRIS_00473 | 470385 | 470951 | 567 | + | PROTEIN:protein kinase |
| LCRIS_00474 | LCRIS_00474 | 470948 | 472567 | 1620 | + | PROTEIN:ABC transporter |
| LCRIS_00475 | pckA | 472656 | 474314 | 1659 | - | PROTEIN:phosphoenolpyruvate carboxykinase (ATP) |
| LCRIS_00476 | LCRIS_00476 | 474527 | 476347 | 1821 | + | PROTEIN:hypothetical protein |
| LCRIS_00477 | LCRIS_00477 | 476352 | 477680 | 1329 | + | PROTEIN:biotin carboxylase |
| LCRIS_00478 | lhr | 477690 | 479927 | 2238 | + | PROTEIN:ATP-dependent helicase |
| LCRIS_00479 | LCRIS_00479 | 479973 | 480344 | 372 | + | PROTEIN:hypothetical protein |
| LCRIS_00480 | LCRIS_00480 | 480469 | 481227 | 759 | + | PROTEIN:nitro/flavin reductase |
| LCRIS_02076 | tRNA-Thr | 481302 | 481377 | 76 | + | RNA:Thr tRNA |
| LCRIS_00481 | LCRIS_00481 | 481596 | 482003 | 408 | - | PROTEIN:hypothetical protein |
| LCRIS_00482 | maa2 | 482144 | 482746 | 603 | + | PROTEIN:maltose o-acetyltransferase |
| LCRIS_00483 | LCRIS_00483 | 482798 | 483193 | 396 | - | PROTEIN:hypothetical protein |
| LCRIS_00484 | LCRIS_00484 | 483290 | 483520 | 231 | + | PROTEIN:hypothetical protein |
| LCRIS_00485 | LCRIS_00485 | 483567 | 483767 | 201 | - | PROTEIN:hypothetical protein |
| LCRIS_00486 | pphA | 483911 | 484783 | 873 | + | PROTEIN:phosphoesterase |
| LCRIS_00487 | LCRIS_00487 | 485253 | 485879 | 627 | + | PROTEIN:resolvase, n terminal domain family protei |
| LCRIS_00488 | LCRIS_00488 | 485884 | 487203 | 1320 | + | PROTEIN:transposase |
| LCRIS_00489 | celB1 | 487397 | 488038 | 642 | + | PROTEIN:PTS system cellobiose-specific iic compone |
| LCRIS_00490 | LCRIS_00490 | 488010 | 488213 | 204 | + | PROTEIN:hypothetical protein |
| LCRIS_00491 | LCRIS_00491 | 488581 | 489252 | 672 | + | PROTEIN:aggregation promoting factor |
| LCRIS_00492 | LCRIS_00492 | 489414 | 490478 | 1065 | + | PROTEIN:surface exclusion protein |
| LCRIS_00493 | gpmB | 490520 | 491095 | 576 | + | PROTEIN:phosphoglycerate mutase |
| LCRIS_00494 | pgm1 | 491097 | 491687 | 591 | + | PROTEIN:phosphoglycerate mutase |
| LCRIS_00495 | LCRIS_00495 | 491756 | 492415 | 660 | + | PROTEIN:hypothetical protein |
| LCRIS_00496 | glpQ2 | 492462 | 493142 | 681 | + | PROTEIN:glycerophosphoryl diester phosphodiesteras |
| LCRIS_00497 | lacI | 493176 | 494147 | 972 | + | PROTEIN:sugar-binding transcriptional regulator, l |
| LCRIS_00498 | LCRIS_00498 | 494344 | 495642 | 1299 | + | PROTEIN:ABC transporter, sugar-binding protein |
| LCRIS_00499 | LCRIS_00499 | 495663 | 496547 | 885 | + | PROTEIN:sugar ABC transporter, permease protein |
| LCRIS_00500 | LCRIS_00500 | 496540 | 497397 | 858 | + | PROTEIN:ABC transporter, permease protein |
| LCRIS_00501 | sacA | 497397 | 498689 | 1293 | + | PROTEIN:sucrose-6-phosphate hydrolase |
| LCRIS_00502 | msmK1 | 498704 | 499810 | 1107 | + | PROTEIN:sugar ABC transporter, ATP-binding protein |
| LCRIS_00503 | gtfA1 | 499932 | 501374 | 1443 | + | PROTEIN:sucrose phosphorylase |
| LCRIS_02077 | tRNA-Gln | 501548 | 501619 | 72 | + | RNA:Gln tRNA |
| LCRIS_00504 | tyrB | 501751 | 502923 | 1173 | + | PROTEIN:aspartate aminotransferase |
| LCRIS_00505 | LCRIS_00505 | 502978 | 503967 | 990 | - | PROTEIN:oxidoreductase family protein |
| LCRIS_00506 | rpiA1 | 503954 | 504637 | 684 | - | PROTEIN:ribose-5-phosphate isomerase |
| LCRIS_00507 | LCRIS_00507 | 504790 | 505623 | 834 | + | PROTEIN:transcriptional regulator |
| LCRIS_00508 | slpX | 505952 | 507427 | 1476 | + | PROTEIN:s-layer protein |
| LCRIS_00509 | LCRIS_00509 | 507572 | 507730 | 159 | + | PROTEIN:hypothetical protein |
| LCRIS_00510 | LCRIS_00510 | 507743 | 507994 | 252 | + | PROTEIN:hypothetical protein |
| LCRIS_00511 | npr | 508254 | 509621 | 1368 | + | PROTEIN:NADH peroxidase |
| LCRIS_02078 | tRNA-Tyr | 509744 | 509825 | 82 | + | RNA:Tyr tRNA |
| LCRIS_02079 | tRNA-Gln | 509860 | 509931 | 72 | + | RNA:Gln tRNA |
| LCRIS_00512 | LCRIS_00512 | 509937 | 511652 | 1716 | - | PROTEIN:transposase |
| LCRIS_00513 | LCRIS_00513 | 511957 | 513540 | 1584 | + | PROTEIN:oligopeptide ABC transporter, oligopeptide |
| LCRIS_00514 | LCRIS_00514 | 513717 | 514766 | 1050 | + | PROTEIN:hypothetical protein |
| LCRIS_00515 | gntR1 | 514854 | 515555 | 702 | + | PROTEIN:transcriptional regulator, gntr family |
| LCRIS_00516 | tagD | 515603 | 515989 | 387 | - | PROTEIN:glycerol-3-phosphate cytidylyltransferase |
| LCRIS_00517 | tagA | 516116 | 516847 | 732 | + | PROTEIN:UDP-N-acetyl-d-mannosamine transferase |
| LCRIS_00518 | LCRIS_00518 | 516858 | 517949 | 1092 | + | PROTEIN:glycosyl transferase, group 1 family prote |
| LCRIS_00519 | pncB | 518036 | 519514 | 1479 | + | PROTEIN:nicotinate phosphoribosyltransferase |
| LCRIS_00520 | nadE | 519511 | 520341 | 831 | + | PROTEIN:nh(3)-dependent nad( ) synthetase |
| LCRIS_00521 | LCRIS_00521 | 520528 | 523197 | 2670 | + | PROTEIN:cation-transporting p-type atpase |
| LCRIS_00522 | LCRIS_00522 | 523287 | 524441 | 1155 | + | PROTEIN:teichoic acid biosynthesis protein |
| LCRIS_00523 | LCRIS_00523 | 524443 | 525873 | 1431 | + | PROTEIN:capsular polysaccharide synthesis protein |
| LCRIS_00524 | LCRIS_00524 | 525876 | 526574 | 699 | + | PROTEIN:glycosyltransferase |
| LCRIS_00525 | sprL | 526634 | 527092 | 459 | + | PROTEIN:sprt-like protein |
| LCRIS_02080 | tRNA-Leu | 527169 | 527253 | 85 | + | RNA:Leu tRNA |
| LCRIS_00526 | LCRIS_00526 | 527339 | 527986 | 648 | + | PROTEIN:N-acetylmuramidase |
| LCRIS_00527 | pcrA | 528072 | 530318 | 2247 | + | PROTEIN:ATP-dependent DNA helicase pcra |
| LCRIS_00528 | ligA | 530353 | 532359 | 2007 | + | PROTEIN:DNA ligase |
| LCRIS_00529 | LCRIS_00529 | 532372 | 533520 | 1149 | + | PROTEIN:sex pheromone biosynthesis protein |
| LCRIS_00530 | gatC | 533532 | 533840 | 309 | + | PROTEIN:aspartyl/glutamyl-tRNA(asn/gln) amidotrans |
| LCRIS_00531 | gatA | 533840 | 535279 | 1440 | + | PROTEIN:aspartyl/glutamyl-tRNA(asn/gln) amidotrans |
| LCRIS_00532 | gatB | 535284 | 536714 | 1431 | + | PROTEIN:aspartyl/glutamyl-tRNA(asn/gln) amidotrans |
| LCRIS_00533 | dagK | 536739 | 537659 | 921 | + | PROTEIN:diacylglycerol kinase |
| LCRIS_00534 | LCRIS_00534 | 537729 | 538439 | 711 | - | PROTEIN:hypothetical protein |
| LCRIS_00535 | LCRIS_00535 | 538964 | 539062 | 99 | + | PROTEIN:hypothetical protein |
| LCRIS_00536 | LCRIS_00536 | 539509 | 540426 | 918 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00537 | LCRIS_00537 | 540413 | 542029 | 1617 | + | PROTEIN:exporter of polyketide antibiotics |
| LCRIS_00538 | rutR | 542026 | 542664 | 639 | + | PROTEIN:transcriptional regulator, tetr family |
| LCRIS_00539 | tnpR | 542781 | 543368 | 588 | + | PROTEIN:resolvase |
| LCRIS_00540 | LCRIS_00540 | 543441 | 544157 | 717 | + | PROTEIN:phage mu protein f like protein |
| LCRIS_00541 | LCRIS_00541 | 544276 | 544524 | 249 | - | PROTEIN:transglycosylase-associated protein |
| LCRIS_00542 | LCRIS_00542 | 544887 | 545807 | 921 | - | PROTEIN:transposase |
| LCRIS_00543 | LCRIS_00543 | 545952 | 546236 | 285 | + | PROTEIN:hypothetical protein |
| LCRIS_00544 | LCRIS_00544 | 546375 | 546749 | 375 | - | PROTEIN:hypothetical protein |
| LCRIS_00545 | LCRIS_00545 | 546742 | 547065 | 324 | - | PROTEIN:hypothetical protein |
| LCRIS_00546 | LCRIS_00546 | 547215 | 547838 | 624 | + | PROTEIN:transposase orf_a |
| LCRIS_00547 | LCRIS_00547 | 547822 | 549150 | 1329 | + | PROTEIN:transposase orf_b |
| LCRIS_00548 | LCRIS_00548 | 549382 | 549729 | 348 | + | PROTEIN:hypothetical protein |
| LCRIS_00549 | LCRIS_00549 | 549704 | 550372 | 669 | - | PROTEIN:hypothetical protein |
| LCRIS_00550 | LCRIS_00550 | 550442 | 550738 | 297 | - | PROTEIN:hypothetical protein |
| LCRIS_00551 | nhaC | 550823 | 552196 | 1374 | - | PROTEIN:Na+/H+ antiporter |
| LCRIS_00552 | pbp1 | 552597 | 553631 | 1035 | + | PROTEIN:penicillin-binding protein |
| LCRIS_00553 | rumA1 | 553715 | 555064 | 1350 | + | PROTEIN:RNA methyltransferase |
| LCRIS_00554 | LCRIS_00554 | 555067 | 555567 | 501 | + | PROTEIN:acetyltransferase |
| LCRIS_02081 | tRNA-Ser | 555658 | 555747 | 90 | + | RNA:Ser tRNA |
| LCRIS_02082 | tRNA-Asp | 555752 | 555825 | 74 | + | RNA:Asp tRNA |
| LCRIS_00555 | LCRIS_00555 | 555801 | 556022 | 222 | + | PROTEIN:hypothetical protein |
| LCRIS_02083 | tRNA-His | 555832 | 555907 | 76 | + | RNA:His tRNA |
| LCRIS_02084 | tRNA-Val | 555912 | 555984 | 73 | + | RNA:Val tRNA |
| LCRIS_00556 | LCRIS_00556 | 556055 | 557521 | 1467 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00557 | LCRIS_00557 | 557564 | 558076 | 513 | - | PROTEIN:transcriptional regulator |
| LCRIS_00558 | mycA1 | 558182 | 559954 | 1773 | + | PROTEIN:67 kda myosin-cross-reactive antigen famil |
| LCRIS_00559 | LCRIS_00559 | 560314 | 560784 | 471 | + | PROTEIN:acetyltransferase |
| LCRIS_00560 | LCRIS_00560 | 560948 | 561784 | 837 | + | PROTEIN:oxidoreductase, aldo/keto reductase family |
| LCRIS_00561 | LCRIS_00561 | 561865 | 562137 | 273 | + | PROTEIN:hypothetical protein |
| LCRIS_00562 | LCRIS_00562 | 562190 | 563722 | 1533 | - | PROTEIN:transcriptional regulator |
| LCRIS_00563 | uraA | 563909 | 565240 | 1332 | - | PROTEIN:uracil permease |
| LCRIS_00564 | pyrR1 | 565252 | 565779 | 528 | - | PROTEIN:uracil phosphoribosyltransferase / pyrimid |
| LCRIS_00565 | LCRIS_00565 | 566064 | 567533 | 1470 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00566 | mdtG1 | 567569 | 568768 | 1200 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00567 | LCRIS_00567 | 568761 | 569519 | 759 | - | PROTEIN:hypothetical protein |
| LCRIS_00568 | LCRIS_00568 | 569626 | 569898 | 273 | - | PROTEIN:hypothetical protein |
| LCRIS_00569 | LCRIS_00569 | 570063 | 570647 | 585 | - | PROTEIN:hypothetical protein |
| LCRIS_00570 | LCRIS_00570 | 570749 | 571831 | 1083 | + | PROTEIN:aminopeptidase i zinc metalloprotease |
| LCRIS_00571 | LCRIS_00571 | 571925 | 573547 | 1623 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00572 | LCRIS_00572 | 573635 | 574555 | 921 | + | PROTEIN:oxidoreductase, aldo/keto reductase family |
| LCRIS_00573 | LCRIS_00573 | 574650 | 575453 | 804 | + | PROTEIN:hypothetical protein |
| LCRIS_00574 | LCRIS_00574 | 575450 | 576769 | 1320 | - | PROTEIN:amino acid permease |
| LCRIS_00575 | LCRIS_00575 | 576996 | 578327 | 1332 | - | PROTEIN:amino acid permease |
| LCRIS_00576 | LCRIS_00576 | 578488 | 579156 | 669 | + | PROTEIN:glutamine amidotransferase class-i |
| LCRIS_00577 | glnH1 | 579199 | 580017 | 819 | - | PROTEIN:glutamine ABC transporter, glutamine-bindi |
| LCRIS_00578 | LCRIS_00578 | 580195 | 580647 | 453 | + | PROTEIN:transcriptional regulator |
| LCRIS_00579 | LCRIS_00579 | 580634 | 582367 | 1734 | + | PROTEIN:ABC transporter, ATP-binding/permease prot |
| LCRIS_00580 | LCRIS_00580 | 582368 | 584137 | 1770 | + | PROTEIN:ABC transporter, ATP-binding/permease prot |
| LCRIS_00581 | LCRIS_00581 | 584318 | 584896 | 579 | + | PROTEIN:ABC-type cobalt transport system, permease |
| LCRIS_00582 | LCRIS_00582 | 585004 | 586395 | 1392 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00583 | udk | 586460 | 587089 | 630 | - | PROTEIN:uridine kinase |
| LCRIS_00584 | glnQ2 | 587728 | 588468 | 741 | + | PROTEIN:glutamine ABC transporter, ATP-binding pro |
| LCRIS_00585 | glnH2 | 588480 | 589298 | 819 | + | PROTEIN:glutamine ABC transporter, glutamine-bindi |
| LCRIS_00586 | glnM | 589298 | 589939 | 642 | + | PROTEIN:glutamine ABC transporter, permease protei |
| LCRIS_00587 | glnP2 | 589957 | 590610 | 654 | + | PROTEIN:glutamine ABC transporter, permease protei |
| LCRIS_00588 | LCRIS_00588 | 590686 | 591597 | 912 | + | PROTEIN:hypothetical protein |
| LCRIS_00589 | LCRIS_00589 | 591684 | 592298 | 615 | + | PROTEIN:resolvase, n terminal domain family protei |
| LCRIS_00590 | LCRIS_00590 | 592295 | 593578 | 1284 | + | PROTEIN:transposase |
| LCRIS_00591 | LCRIS_00591 | 593838 | 594335 | 498 | - | PROTEIN:hypothetical protein |
| LCRIS_00592 | LCRIS_00592 | 594487 | 595887 | 1401 | + | PROTEIN:hypothetical protein |
| LCRIS_00593 | LCRIS_00593 | 595951 | 597255 | 1305 | - | PROTEIN:glycerol-3-phosphate ABC transporter |
| LCRIS_00594 | LCRIS_00594 | 597419 | 598222 | 804 | + | PROTEIN:galactose-1-phosphate uridylyltransferase |
| LCRIS_00595 | rbsK | 598312 | 599238 | 927 | + | PROTEIN:ribokinase |
| LCRIS_00596 | rpiA2 | 599238 | 599930 | 693 | + | PROTEIN:ribose-5-phosphate isomerase a |
| LCRIS_00597 | kup2 | 600069 | 602078 | 2010 | + | PROTEIN:potassium transport system protein kup |
| LCRIS_00598 | iunH1 | 602160 | 603086 | 927 | + | PROTEIN:inosine-uridine preferring nucleoside hydr |
| LCRIS_00599 | LCRIS_00599 | 603086 | 603811 | 726 | + | PROTEIN:transcriptional regulator, gntr family |
| LCRIS_00600 | LCRIS_00600 | 603905 | 604732 | 828 | - | PROTEIN:hydrolase |
| LCRIS_00601 | LCRIS_00601 | 604854 | 605213 | 360 | - | PROTEIN:hypothetical protein |
| LCRIS_00602 | LCRIS_00602 | 605219 | 605983 | 765 | - | PROTEIN:transcriptional regulator |
| LCRIS_00603 | xfp | 606284 | 608683 | 2400 | + | PROTEIN:xylulose-5-phosphate/fructose-6-phosphate |
| LCRIS_00604 | LCRIS_00604 | 608751 | 609599 | 849 | + | PROTEIN:serine protease |
| LCRIS_00605 | LCRIS_00605 | 609655 | 610461 | 807 | - | PROTEIN:hydrolase |
| LCRIS_00606 | malX | 611071 | 612951 | 1881 | + | PROTEIN:PTS system alpha-glucoside-specific iibc c |
| LCRIS_00607 | glvR | 613013 | 613822 | 810 | + | PROTEIN:transcriptional regulator |
| LCRIS_00608 | LCRIS_00608 | 613809 | 614306 | 498 | + | PROTEIN:hypothetical protein |
| LCRIS_00609 | crr1 | 614327 | 614812 | 486 | + | PROTEIN:PTS system glucose-specific iia component |
| LCRIS_00610 | LCRIS_00610 | 614931 | 615197 | 267 | + | PROTEIN:hypothetical protein |
| LCRIS_00611 | LCRIS_00611 | 615269 | 615925 | 657 | + | PROTEIN:p-nitrobenzoate reductase |
| LCRIS_00612 | LCRIS_00612 | 616028 | 617386 | 1359 | + | PROTEIN:transport protein |
| LCRIS_00613 | hpt1 | 617493 | 618062 | 570 | + | PROTEIN:hypoxanthine phosphoribosyltransferase |
| LCRIS_00614 | frdA1 | 618235 | 620004 | 1770 | + | PROTEIN:fumarate reductase flavoprotein subunit |
| LCRIS_00615 | LCRIS_00615 | 620084 | 620251 | 168 | - | PROTEIN:hypothetical protein |
| LCRIS_00616 | LCRIS_00616 | 620351 | 620626 | 276 | - | PROTEIN:polynucleotidyl transferase, ribonuclease |
| LCRIS_00617 | LCRIS_00617 | 620604 | 620915 | 312 | - | PROTEIN:hypothetical protein |
| LCRIS_00618 | LCRIS_00618 | 621238 | 623118 | 1881 | + | PROTEIN:fumarate reductase flavoprotein subunit |
| LCRIS_00619 | LCRIS_00619 | 623170 | 624471 | 1302 | - | PROTEIN:amino acid permease |
| LCRIS_00620 | LCRIS_00620 | 624580 | 625287 | 708 | + | PROTEIN:lyzozyme m1 (1,4-beta-N-acetylmuramidase) |
| LCRIS_00621 | LCRIS_00621 | 625290 | 626525 | 1236 | + | PROTEIN:ATP-dependent RNA helicase, dead-deah box |
| LCRIS_00622 | celB2 | 626522 | 627850 | 1329 | + | PROTEIN:PTS system cellobiose-specific iic compone |
| LCRIS_02085 | tRNA-Arg | 627900 | 627972 | 73 | + | RNA:Arg tRNA |
| LCRIS_00623 | LCRIS_00623 | 628043 | 628231 | 189 | - | PROTEIN:hypothetical protein |
| LCRIS_00624 | wecB | 628337 | 629479 | 1143 | + | PROTEIN:UDP-N-acetylglucosamine 2-epimerase |
| LCRIS_00625 | LCRIS_00625 | 629443 | 630024 | 582 | - | PROTEIN:cell wall teichoic acid glycosylation prot |
| LCRIS_00626 | LCRIS_00626 | 630017 | 630463 | 447 | - | PROTEIN:flavodoxin |
| LCRIS_00627 | map | 630676 | 631503 | 828 | + | PROTEIN:methionine aminopeptidase |
| LCRIS_00628 | LCRIS_00628 | 631512 | 632435 | 924 | + | PROTEIN:hypothetical protein |
| LCRIS_00629 | galU | 632496 | 633398 | 903 | + | PROTEIN:utp-glucose-1-phosphate uridylyltransferas |
| LCRIS_00630 | atoB | 633509 | 634672 | 1164 | + | PROTEIN:acetyl-CoA acetyltransferase |
| LCRIS_00631 | mvaA | 634669 | 635880 | 1212 | + | PROTEIN:hydroxymethylglutaryl-CoA reductase |
| LCRIS_00632 | pksG | 635880 | 637046 | 1167 | + | PROTEIN:hydroxymethylglutaryl-CoA synthase |
| LCRIS_00633 | LCRIS_00633 | 637074 | 637358 | 285 | - | PROTEIN:hypothetical protein |
| LCRIS_00634 | LCRIS_00634 | 637452 | 638372 | 921 | + | PROTEIN:hydrolase of the alpha/beta superfamily |
| LCRIS_00635 | rumA2 | 638812 | 640200 | 1389 | - | PROTEIN:RNA methyltransferase |
| LCRIS_00636 | recX | 640325 | 641137 | 813 | + | PROTEIN:regulatory protein recx |
| LCRIS_00637 | LCRIS_00637 | 641146 | 641628 | 483 | + | PROTEIN:hypothetical protein |
| LCRIS_00638 | LCRIS_00638 | 641629 | 642090 | 462 | - | PROTEIN:hypothetical protein |
| LCRIS_00639 | tlyC | 642341 | 643060 | 720 | + | PROTEIN:cystathionine beta-synthase |
| LCRIS_00640 | prfC | 643060 | 644631 | 1572 | + | PROTEIN:peptide chain release factor 3 |
| LCRIS_00641 | LCRIS_00641 | 644646 | 645455 | 810 | + | PROTEIN:transcriptional regulator, xre family |
| LCRIS_00642 | LCRIS_00642 | 645557 | 647125 | 1569 | + | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_00643 | LCRIS_00643 | 647249 | 647992 | 744 | + | PROTEIN:hypothetical protein |
| LCRIS_00644 | LCRIS_00644 | 648076 | 648264 | 189 | + | PROTEIN:hypothetical protein |
| LCRIS_00645 | LCRIS_00645 | 648359 | 648523 | 165 | + | PROTEIN:hypothetical protein |
| LCRIS_00646 | LCRIS_00646 | 648635 | 648967 | 333 | + | PROTEIN:cassette chromosome recombinase b1 |
| LCRIS_00647 | LCRIS_00647 | 648979 | 649176 | 198 | + | PROTEIN:transposase (iscpe5) |
| LCRIS_00648 | LCRIS_00648 | 649356 | 649637 | 282 | - | PROTEIN:hypothetical protein |
| LCRIS_00649 | clpE1 | 649783 | 651975 | 2193 | - | PROTEIN:ATP-dependent clp protease, ATP-binding su |
| LCRIS_00650 | LCRIS_00650 | 652180 | 652362 | 183 | + | PROTEIN:hypothetical protein |
| LCRIS_00651 | ptsH | 652475 | 652741 | 267 | + | PROTEIN:phosphocarrier protein hpr |
| LCRIS_00652 | ptsI | 652741 | 654474 | 1734 | + | PROTEIN:phosphoenolpyruvate-protein phosphotransfe |
| LCRIS_00653 | arsC | 654698 | 655096 | 399 | + | PROTEIN:arsenate reductase related protein, glutar |
| LCRIS_00654 | mecA | 655201 | 655947 | 747 | + | PROTEIN:negative regulator genetic competence |
| LCRIS_00655 | coiA | 656019 | 656867 | 849 | + | PROTEIN:competence protein |
| LCRIS_00656 | LCRIS_00656 | 656875 | 657492 | 618 | - | PROTEIN:dithiol-disulfide isomerase |
| LCRIS_00657 | LCRIS_00657 | 657579 | 658187 | 609 | - | PROTEIN:adenylate cyclase family |
| LCRIS_00658 | LCRIS_00658 | 658264 | 658896 | 633 | + | PROTEIN:GTP pyrophosphokinase |
| LCRIS_00659 | ppnK | 658893 | 659696 | 804 | + | PROTEIN:inorganic polyphosphate/ATP-nad kinase |
| LCRIS_00660 | rluD1 | 659689 | 660594 | 906 | + | PROTEIN:pseudouridine synthase |
| LCRIS_00661 | mycA2 | 660700 | 662475 | 1776 | + | PROTEIN:67 kda myosin-cross-reactive antigen famil |
| LCRIS_00662 | LCRIS_00662 | 662510 | 663055 | 546 | - | PROTEIN:hypothetical protein |
| LCRIS_00663 | pgl | 663314 | 664342 | 1029 | - | PROTEIN:3-carboxymuconate cyclase |
| LCRIS_00664 | srlB | 664522 | 664902 | 381 | + | PROTEIN:PTS system glucitol/sorbitol-specific iia |
| LCRIS_00665 | LCRIS_00665 | 664919 | 666106 | 1188 | + | PROTEIN:permease |
| LCRIS_00666 | cspR | 666169 | 666729 | 561 | + | PROTEIN:RNA methyltransferase, trmh family, group |
| LCRIS_00667 | LCRIS_00667 | 666729 | 667124 | 396 | + | PROTEIN:hypothetical protein |
| LCRIS_00668 | ftsK | 667137 | 669560 | 2424 | + | PROTEIN:DNA translocase ftsk |
| LCRIS_00669 | LCRIS_00669 | 669547 | 670761 | 1215 | + | PROTEIN:protease |
| LCRIS_00670 | LCRIS_00670 | 670758 | 672002 | 1245 | + | PROTEIN:protease |
| LCRIS_00671 | fabG | 672016 | 672744 | 729 | + | PROTEIN:3-oxoacyl-(acyl-carrier protein) reductase |
| LCRIS_00672 | LCRIS_00672 | 672812 | 673891 | 1080 | + | PROTEIN:transcriptional regulator, xre family |
| LCRIS_00673 | pgsA | 673909 | 674469 | 561 | + | PROTEIN:cdp-diacylglycerol--glycerol-3-phosphate 3 |
| LCRIS_00675 | LCRIS_00675 | 674482 | 674667 | 186 | - | PROTEIN:hypothetical protein |
| LCRIS_00674 | recA | 674660 | 675760 | 1101 | + | PROTEIN:protein reca |
| LCRIS_00676 | ymdA | 675876 | 677513 | 1638 | + | PROTEIN:2',3'-cyclic-nucleotide 2'-phosphodiestera |
| LCRIS_00677 | rfe | 677618 | 678775 | 1158 | + | PROTEIN:glycosyl transferase, group 4 family prote |
| LCRIS_00678 | yvyE | 678810 | 679472 | 663 | - | PROTEIN:yvye |
| LCRIS_00679 | comFA | 679518 | 680798 | 1281 | + | PROTEIN:comf operon protein 1 |
| LCRIS_00680 | comFC | 680795 | 681490 | 696 | + | PROTEIN:competence protein |
| LCRIS_00681 | yfiA | 681571 | 682116 | 546 | + | PROTEIN:ribosomal subunit interface protein |
| LCRIS_00682 | secA | 682258 | 684657 | 2400 | + | PROTEIN:protein translocase subunit seca |
| LCRIS_00683 | prfB | 684840 | 685838 | 999 | + | PROTEIN:peptide chain release factor 2 |
| LCRIS_00684 | LCRIS_00684 | 685847 | 686119 | 273 | + | PROTEIN:hypothetical protein |
| LCRIS_00685 | ptsK | 686134 | 687102 | 969 | + | PROTEIN:hpr kinase/phosphorylase |
| LCRIS_00686 | lgt | 687095 | 687934 | 840 | + | PROTEIN:prolipoprotein diacylglyceryl transferase |
| LCRIS_00687 | gpsA | 687975 | 688994 | 1020 | + | PROTEIN:glycerol-3-phosphate dehydrogenase [nad(p) |
| LCRIS_00688 | trxB2 | 689076 | 690014 | 939 | + | PROTEIN:thioredoxin reductase |
| LCRIS_00689 | pgm2 | 690181 | 691905 | 1725 | + | PROTEIN:phosphoglucomutase |
| LCRIS_00690 | uvrB | 692008 | 694053 | 2046 | + | PROTEIN:uvrABC system protein b |
| LCRIS_00691 | uvrA1 | 694043 | 696883 | 2841 | + | PROTEIN:uvrABC system protein a |
| LCRIS_00692 | LCRIS_00692 | 696956 | 697498 | 543 | + | PROTEIN:hypothetical protein |
| LCRIS_00693 | LCRIS_00693 | 697592 | 698473 | 882 | + | PROTEIN:P-loop ATPase |
| LCRIS_00694 | LCRIS_00694 | 698484 | 699527 | 1044 | + | PROTEIN:transporter |
| LCRIS_00695 | whiA | 699520 | 700455 | 936 | + | PROTEIN:sporulation transcription regulator whia |
| LCRIS_00696 | clpP | 700520 | 701104 | 585 | - | PROTEIN:ATP-dependent clp protease proteolytic sub |
| LCRIS_00697 | LCRIS_00697 | 701299 | 702927 | 1629 | + | PROTEIN:conserved protein with bacterial ig-like d |
| LCRIS_00698 | ybbL | 703036 | 703674 | 639 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00699 | ybbM | 703671 | 704429 | 759 | + | PROTEIN:ABC-type transport system, permease compon |
| LCRIS_00700 | LCRIS_00700 | 704676 | 705236 | 561 | - | PROTEIN:hypothetical protein |
| LCRIS_00701 | LCRIS_00701 | 705237 | 706709 | 1473 | - | PROTEIN:drug resistance transporter, emrb/qaca fam |
| LCRIS_00702 | LCRIS_00702 | 706870 | 707307 | 438 | + | PROTEIN:transcriptional regulator, merr family |
| LCRIS_00703 | LCRIS_00703 | 707391 | 708176 | 786 | + | PROTEIN:hypothetical protein |
| LCRIS_00704 | LCRIS_00704 | 708243 | 709676 | 1434 | - | PROTEIN:amino acid permease |
| LCRIS_00705 | LCRIS_00705 | 709845 | 711377 | 1533 | + | PROTEIN:conserved protein with bacterial ig-like d |
| LCRIS_02097 | tRNA-Arg | 711472 | 711543 | 72 | - | RNA:Arg tRNA |
| LCRIS_00706 | cggR | 711791 | 712822 | 1032 | + | PROTEIN:central glycolytic genes regulator |
| LCRIS_00707 | gapA | 712875 | 713891 | 1017 | + | PROTEIN:glyceraldehyde-3-phosphate dehydrogenase |
| LCRIS_00708 | pgk | 714006 | 715217 | 1212 | + | PROTEIN:phosphoglycerate kinase |
| LCRIS_00709 | tpiA | 715242 | 716000 | 759 | + | PROTEIN:triosephosphate isomerase |
| LCRIS_00710 | LCRIS_00710 | 716487 | 717002 | 516 | - | PROTEIN:hypothetical protein |
| LCRIS_00711 | LCRIS_00711 | 717022 | 717897 | 876 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_00712 | ung | 717965 | 718663 | 699 | + | PROTEIN:uracil-DNA glycosylase |
| LCRIS_00713 | pta | 718674 | 719663 | 990 | + | PROTEIN:phosphate acetyltransferase |
| LCRIS_00714 | LCRIS_00714 | 719663 | 720142 | 480 | + | PROTEIN:atpase or kinase |
| LCRIS_00715 | dnaQ1 | 720180 | 720713 | 534 | - | PROTEIN:DNA polymerase iii, epsilon subunit relate |
| LCRIS_00716 | murB | 720809 | 721705 | 897 | + | PROTEIN:UDP-N-acetylenolpyruvoylglucosamine reduct |
| LCRIS_00717 | potA | 721772 | 722869 | 1098 | + | PROTEIN:spermidine/putrescine ABC transporter, ATP |
| LCRIS_00718 | potB | 722859 | 723671 | 813 | + | PROTEIN:spermidine/putrescine ABC transporter, per |
| LCRIS_00719 | potC | 723668 | 724483 | 816 | + | PROTEIN:spermidine/putrescine ABC transporter, per |
| LCRIS_00720 | potD | 724480 | 725553 | 1074 | + | PROTEIN:spermidine/putrescine ABC transporter, spe |
| LCRIS_00721 | LCRIS_00721 | 725592 | 726434 | 843 | + | PROTEIN:hypothetical protein |
| LCRIS_00722 | ybbR | 726431 | 727390 | 960 | + | PROTEIN:ybbr |
| LCRIS_00723 | glmM | 727413 | 728765 | 1353 | + | PROTEIN:phosphoglucosamine mutase |
| LCRIS_00724 | LCRIS_00724 | 728866 | 729681 | 816 | + | PROTEIN:had superfamily hydrolase |
| LCRIS_00725 | copY | 729712 | 730152 | 441 | + | PROTEIN:transcriptional repressor copy |
| LCRIS_00726 | LCRIS_00726 | 730167 | 730883 | 717 | + | PROTEIN:hypothetical protein |
| LCRIS_00727 | ptpA | 730883 | 731338 | 456 | + | PROTEIN:phosphotyrosine protein phosphatase |
| LCRIS_00728 | glxK | 731363 | 732508 | 1146 | - | PROTEIN:glycerate kinase |
| LCRIS_00729 | bglG1 | 732717 | 733550 | 834 | + | PROTEIN:transcription antiterminator |
| LCRIS_00730 | bglF1 | 733678 | 735699 | 2022 | + | PROTEIN:PTS system beta-glucoside-specific iiABC s |
| LCRIS_00731 | bglA1 | 735741 | 737216 | 1476 | + | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_00732 | LCRIS_00732 | 737388 | 738338 | 951 | + | PROTEIN:hydrolase of the alpha/beta superfamily |
| LCRIS_00733 | LCRIS_00733 | 738383 | 740008 | 1626 | - | PROTEIN:amino acid transporter |
| LCRIS_00734 | rpsN | 740384 | 740569 | 186 | + | PROTEIN:30S ribosomal protein s14 type z |
| LCRIS_00735 | LCRIS_00735 | 740547 | 741173 | 627 | + | PROTEIN:nucleoside-diphosphate-sugar epimerase |
| LCRIS_00736 | LCRIS_00736 | 741245 | 741766 | 522 | + | PROTEIN:integral hypothetical protein |
| LCRIS_00737 | LCRIS_00737 | 741851 | 742579 | 729 | + | PROTEIN:hypothetical protein |
| LCRIS_00738 | comGA | 742777 | 743751 | 975 | + | PROTEIN:competence protein |
| LCRIS_00739 | comGB | 743720 | 744721 | 1002 | + | PROTEIN:competence protein |
| LCRIS_00740 | comGC | 744781 | 745122 | 342 | + | PROTEIN:competence protein |
| LCRIS_00741 | comGD | 745109 | 745522 | 414 | + | PROTEIN:prepilin-type cleavage/methylation protein |
| LCRIS_00742 | LCRIS_00742 | 745519 | 745788 | 270 | + | PROTEIN:hypothetical protein |
| LCRIS_00743 | LCRIS_00743 | 745796 | 746269 | 474 | + | PROTEIN:hypothetical protein |
| LCRIS_00744 | LCRIS_00744 | 746241 | 746405 | 165 | + | PROTEIN:hypothetical protein |
| LCRIS_00745 | LCRIS_00745 | 746464 | 747465 | 1002 | + | PROTEIN:adenine-specific DNA methylase |
| LCRIS_00746 | ackA1 | 747509 | 748693 | 1185 | + | PROTEIN:acetate kinase |
| LCRIS_00747 | LCRIS_00747 | 749501 | 750214 | 714 | + | PROTEIN:hypothetical protein |
| LCRIS_00748 | patB | 750223 | 751386 | 1164 | + | PROTEIN:aminotransferase |
| LCRIS_00749 | manA | 751479 | 752441 | 963 | + | PROTEIN:mannose-6-phosphate isomerase |
| LCRIS_00750 | LCRIS_00750 | 752477 | 752866 | 390 | + | PROTEIN:integral hypothetical protein |
| LCRIS_00751 | LCRIS_00751 | 752869 | 753591 | 723 | + | PROTEIN:response regulator |
| LCRIS_00752 | LCRIS_00752 | 753591 | 755045 | 1455 | + | PROTEIN:sensor protein |
| LCRIS_00753 | LCRIS_00753 | 755089 | 757041 | 1953 | - | PROTEIN:sulfatase |
| LCRIS_00754 | LCRIS_00754 | 757411 | 758013 | 603 | + | PROTEIN:hypothetical protein |
| LCRIS_00755 | pgi | 758101 | 759438 | 1338 | + | PROTEIN:glucose-6-phosphate isomerase |
| LCRIS_00756 | LCRIS_00756 | 759557 | 760753 | 1197 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00757 | LCRIS_00757 | 760708 | 760929 | 222 | + | PROTEIN:hypothetical protein |
| LCRIS_00758 | LCRIS_00758 | 761038 | 762048 | 1011 | - | PROTEIN:beta-lactamase class c related penicillin |
| LCRIS_02086 | tRNA-Glu | 762130 | 762201 | 72 | + | RNA:Glu tRNA |
| LCRIS_02087 | tRNA-Gln | 762207 | 762279 | 73 | + | RNA:Gln tRNA |
| LCRIS_00759 | murE1 | 762357 | 763709 | 1353 | - | PROTEIN:UDP-N-acetylmuramyl tripeptide synthetase |
| LCRIS_00760 | tdk | 763845 | 764444 | 600 | + | PROTEIN:thymidine kinase |
| LCRIS_00761 | prfA | 764461 | 765549 | 1089 | + | PROTEIN:peptide chain release factor 1 |
| LCRIS_00762 | hemK | 765542 | 766387 | 846 | + | PROTEIN:protoporphyrinogen oxidase |
| LCRIS_00763 | sua | 766393 | 767394 | 1002 | + | PROTEIN:sua5/ycio/yrdc/ywlc family protein |
| LCRIS_00764 | upp | 767479 | 768108 | 630 | + | PROTEIN:uracil phosphoribosyltransferase |
| LCRIS_00765 | atpB | 768231 | 768944 | 714 | + | PROTEIN:ATP synthase subunit a |
| LCRIS_00766 | atpE | 768965 | 769189 | 225 | + | PROTEIN:ATP synthase subunit c |
| LCRIS_00767 | atpF | 769246 | 769752 | 507 | + | PROTEIN:ATP synthase subunit b |
| LCRIS_00768 | atpH | 769752 | 770300 | 549 | + | PROTEIN:ATP synthase subunit delta |
| LCRIS_00769 | atpA | 770306 | 771826 | 1521 | + | PROTEIN:ATP synthase subunit alpha |
| LCRIS_00770 | atpG | 771837 | 772799 | 963 | + | PROTEIN:ATP synthase subunit gamma |
| LCRIS_00771 | atpD | 772822 | 774261 | 1440 | + | PROTEIN:ATP synthase subunit beta |
| LCRIS_00772 | atpC | 774273 | 774713 | 441 | + | PROTEIN:ATP synthase subunit epsilon |
| LCRIS_00773 | LCRIS_00773 | 774780 | 775010 | 231 | + | PROTEIN:hypothetical protein |
| LCRIS_00774 | mreB1 | 775094 | 776083 | 990 | + | PROTEIN:rod shape-determining protein mreb |
| LCRIS_00775 | LCRIS_00775 | 776097 | 776390 | 294 | + | PROTEIN:hypothetical protein |
| LCRIS_00776 | LCRIS_00776 | 776399 | 776626 | 228 | + | PROTEIN:hypothetical protein |
| LCRIS_00777 | rodA | 776648 | 777841 | 1194 | + | PROTEIN:cell division hypothetical protein |
| LCRIS_00778 | LCRIS_00778 | 777922 | 778386 | 465 | - | PROTEIN:universal stress protein family |
| LCRIS_00779 | LCRIS_00779 | 778447 | 778728 | 282 | - | PROTEIN:hypothetical protein |
| LCRIS_00780 | ycaJ | 779039 | 780346 | 1308 | - | PROTEIN:atpase, aaa family |
| LCRIS_00781 | yueI | 780347 | 780814 | 468 | - | PROTEIN:yuei protein |
| LCRIS_00782 | rpsD | 780906 | 781517 | 612 | - | PROTEIN:30S ribosomal protein s4 |
| LCRIS_00783 | ezrA | 781802 | 783514 | 1713 | + | PROTEIN:negative regulator of septation ring forma |
| LCRIS_00784 | nifS | 783602 | 784762 | 1161 | + | PROTEIN:aminotransferase class v |
| LCRIS_00785 | thiI | 784762 | 785979 | 1218 | + | PROTEIN:thiamine biosynthesis protein thii |
| LCRIS_00786 | rsuA1 | 786026 | 786736 | 711 | + | PROTEIN:pseudouridine synthase |
| LCRIS_00787 | LCRIS_00787 | 786878 | 787687 | 810 | + | PROTEIN:transcriptional regulator |
| LCRIS_00788 | LCRIS_00788 | 787684 | 788433 | 750 | + | PROTEIN:regulatory protein munl |
| LCRIS_00789 | LCRIS_00789 | 788532 | 790256 | 1725 | + | PROTEIN:hypothetical protein |
| LCRIS_00790 | LCRIS_00790 | 790550 | 791374 | 825 | + | PROTEIN:transcriptional regulator |
| LCRIS_00791 | LCRIS_00791 | 791459 | 791986 | 528 | + | PROTEIN:hypothetical protein |
| LCRIS_00792 | LCRIS_00792 | 792279 | 795185 | 2907 | + | PROTEIN:hypothetical protein |
| LCRIS_00793 | LCRIS_00793 | 795315 | 796445 | 1131 | + | PROTEIN:hypothetical protein |
| LCRIS_00794 | LCRIS_00794 | 796711 | 796938 | 228 | - | PROTEIN:transposase |
| LCRIS_00795 | LCRIS_00795 | 797002 | 797991 | 990 | - | PROTEIN:transposase |
| LCRIS_00796 | LCRIS_00796 | 798244 | 801111 | 2868 | + | PROTEIN:hypothetical protein |
| LCRIS_00797 | LCRIS_00797 | 801437 | 802027 | 591 | + | PROTEIN:hypothetical protein |
| LCRIS_00798 | LCRIS_00798 | 802046 | 802306 | 261 | + | PROTEIN:hth-type transcriptional regulator yqaf |
| LCRIS_00799 | LCRIS_00799 | 802459 | 802854 | 396 | + | PROTEIN:hypothetical protein |
| LCRIS_00800 | LCRIS_00800 | 802873 | 803895 | 1023 | + | PROTEIN:hypothetical protein |
| LCRIS_00801 | rsuA2 | 804040 | 804744 | 705 | + | PROTEIN:pseudouridine synthase |
| LCRIS_00802 | LCRIS_00802 | 804845 | 805726 | 882 | + | PROTEIN:hypothetical protein |
| LCRIS_00803 | LCRIS_00803 | 805911 | 806330 | 420 | + | PROTEIN:dolichyl-phosphate beta-d-mannosyltransfer |
| LCRIS_00804 | LCRIS_00804 | 806330 | 807259 | 930 | + | PROTEIN:bactoprenol glucosyl transferase |
| LCRIS_00805 | LCRIS_00805 | 807407 | 807919 | 513 | + | PROTEIN:hypothetical protein |
| LCRIS_00806 | LCRIS_00806 | 807916 | 808191 | 276 | + | PROTEIN:hypothetical protein |
| LCRIS_00807 | LCRIS_00807 | 808225 | 808431 | 207 | + | PROTEIN:hypothetical protein |
| LCRIS_00808 | LCRIS_00808 | 808637 | 811252 | 2616 | + | PROTEIN:cation-transporting atpase |
| LCRIS_00809 | LCRIS_00809 | 811345 | 812274 | 930 | + | PROTEIN:hypothetical protein |
| LCRIS_00810 | LCRIS_00810 | 812325 | 813212 | 888 | - | PROTEIN:transcriptional regulator |
| LCRIS_00811 | LCRIS_00811 | 813807 | 814172 | 366 | + | PROTEIN:hypothetical protein |
| LCRIS_00812 | LCRIS_00812 | 814172 | 814441 | 270 | + | PROTEIN:transcriptional regulator |
| LCRIS_00813 | valS | 814753 | 817392 | 2640 | + | PROTEIN:valyl-tRNA synthetase |
| LCRIS_00814 | folC | 817393 | 818676 | 1284 | + | PROTEIN:folylpolyglutamate synthase |
| LCRIS_00815 | LCRIS_00815 | 818654 | 819331 | 678 | + | PROTEIN:hydrolase |
| LCRIS_00816 | radC | 819367 | 819999 | 633 | + | PROTEIN:DNA repair protein radc |
| LCRIS_00817 | mreB2 | 820085 | 821089 | 1005 | + | PROTEIN:rod shape-determining protein mreb |
| LCRIS_00818 | mreC | 821135 | 821986 | 852 | + | PROTEIN:rod shape-determining protein mrec |
| LCRIS_00819 | mreD | 821986 | 822525 | 540 | + | PROTEIN:rod shape-determining protein mred |
| LCRIS_00820 | LCRIS_00820 | 822786 | 823115 | 330 | + | PROTEIN:hypothetical protein |
| LCRIS_00821 | mraZ | 823286 | 823717 | 432 | + | PROTEIN:protein mraz |
| LCRIS_00822 | mraW | 823723 | 824670 | 948 | + | PROTEIN:s-adenosyl-l-methionine-dependent methyltr |
| LCRIS_00823 | ftsL | 824684 | 825046 | 363 | + | PROTEIN:cell division protein ftsl |
| LCRIS_00824 | pbpB | 825124 | 827205 | 2082 | + | PROTEIN:penicillin-binding protein |
| LCRIS_00825 | mraY | 827212 | 828180 | 969 | + | PROTEIN:phospho-N-acetylmuramoyl-pentapeptide-tran |
| LCRIS_00826 | murD | 828188 | 829567 | 1380 | + | PROTEIN:UDP-N-acetylmuramoylalanine--d-glutamate l |
| LCRIS_00827 | murG | 829569 | 830675 | 1107 | + | PROTEIN:UDP-N-acetylglucosamine--N-acetylmuramyl-( |
| LCRIS_00828 | ftsQ | 830692 | 831549 | 858 | + | PROTEIN:cell division protein ftsq |
| LCRIS_00829 | ftsA | 831612 | 832973 | 1362 | + | PROTEIN:cell division protein ftsa |
| LCRIS_00830 | ftsZ | 832988 | 834331 | 1344 | + | PROTEIN:cell division protein ftsz |
| LCRIS_00831 | sepF | 834349 | 834786 | 438 | + | PROTEIN:cell division protein sepf |
| LCRIS_00832 | yggT | 834786 | 835088 | 303 | + | PROTEIN:cell division hypothetical protein |
| LCRIS_00833 | LCRIS_00833 | 835088 | 835888 | 801 | + | PROTEIN:cell division protein, s4-like domain |
| LCRIS_00834 | divIVA | 835864 | 836688 | 825 | + | PROTEIN:cell division initiation protein |
| LCRIS_00835 | ileS | 836916 | 839702 | 2787 | + | PROTEIN:isoleucyl-tRNA synthetase |
| LCRIS_00836 | cspA | 839702 | 839905 | 204 | + | PROTEIN:cold shock protein |
| LCRIS_00837 | nudF | 839921 | 840490 | 570 | + | PROTEIN:ADP-ribose pyrophosphatase |
| LCRIS_00838 | LCRIS_00838 | 840493 | 840807 | 315 | + | PROTEIN:hypothetical protein |
| LCRIS_00839 | mtnN | 840826 | 841521 | 696 | + | PROTEIN:5'-methylthioadenosine/s-adenosylhomocyste |
| LCRIS_00840 | iscS | 841524 | 842681 | 1158 | + | PROTEIN:cysteine desulfurase |
| LCRIS_00841 | LCRIS_00841 | 842729 | 843070 | 342 | + | PROTEIN:hypothetical protein |
| LCRIS_00842 | mnmA | 843078 | 844205 | 1128 | + | PROTEIN:tRNA-specific 2-thiouridylase mnma |
| LCRIS_00843 | gpm2 | 844298 | 844957 | 660 | + | PROTEIN:phosphoglycerate mutase |
| LCRIS_00844 | LCRIS_00844 | 844957 | 845607 | 651 | + | PROTEIN:tpr repeat protein |
| LCRIS_00845 | recD | 845600 | 847969 | 2370 | + | PROTEIN:exodeoxyribonuclease v alpha chain |
| LCRIS_00846 | LCRIS_00846 | 848151 | 849713 | 1563 | + | PROTEIN:hypothetical protein |
| LCRIS_00847 | LCRIS_00847 | 849739 | 850686 | 948 | - | PROTEIN:diacylglycerol kinase family protein |
| LCRIS_00848 | LCRIS_00848 | 850733 | 852412 | 1680 | - | PROTEIN:metallo-beta-lactamase superfamily protein |
| LCRIS_00849 | LCRIS_00849 | 852419 | 852640 | 222 | - | PROTEIN:hypothetical protein |
| LCRIS_00850 | LCRIS_00850 | 852764 | 852928 | 165 | - | PROTEIN:hypothetical protein |
| LCRIS_00851 | def2 | 852971 | 853525 | 555 | - | PROTEIN:peptide deformylase |
| LCRIS_00852 | typA | 853778 | 855622 | 1845 | + | PROTEIN:GTP-binding protein typa |
| LCRIS_00853 | ftsW | 855722 | 856906 | 1185 | + | PROTEIN:cell division protein ftsw |
| LCRIS_00854 | LCRIS_00854 | 856903 | 857247 | 345 | + | PROTEIN:hypothetical protein |
| LCRIS_00855 | rsmD | 857244 | 857792 | 549 | + | PROTEIN:methyltransferase |
| LCRIS_00856 | coaD | 857795 | 858280 | 486 | + | PROTEIN:phosphopantetheine adenylyltransferase |
| LCRIS_00857 | LCRIS_00857 | 858273 | 859316 | 1044 | + | PROTEIN:ATP-dependent protease la |
| LCRIS_00858 | comEA | 859392 | 860081 | 690 | + | PROTEIN:competence protein comea |
| LCRIS_00859 | comEC | 860050 | 862338 | 2289 | + | PROTEIN:competence protein comec |
| LCRIS_00860 | holA | 862335 | 863321 | 987 | + | PROTEIN:DNA polymerase iii, delta subunit |
| LCRIS_00861 | rpsT | 863385 | 863642 | 258 | - | PROTEIN:30S ribosomal protein s20 |
| LCRIS_00862 | rpsO | 863844 | 864113 | 270 | + | PROTEIN:30S ribosomal protein s15 |
| LCRIS_00863 | LCRIS_00863 | 864271 | 866061 | 1791 | + | PROTEIN:metallo-beta-lactamase superfamily protein |
| LCRIS_00864 | LCRIS_00864 | 866064 | 866897 | 834 | + | PROTEIN:bs_ysoa-like protein with tpr repeats |
| LCRIS_00865 | tufA | 867086 | 868276 | 1191 | + | PROTEIN:elongation factor tu |
| LCRIS_00866 | tig | 868422 | 869753 | 1332 | + | PROTEIN:trigger factor |
| LCRIS_00867 | clpX | 869882 | 871147 | 1266 | + | PROTEIN:ATP-dependent clp protease ATP-binding sub |
| LCRIS_00868 | engB | 871137 | 871727 | 591 | + | PROTEIN:GTP-binding protein engb |
| LCRIS_00869 | LCRIS_00869 | 871853 | 872041 | 189 | + | PROTEIN:hypothetical protein |
| LCRIS_00870 | LCRIS_00870 | 872168 | 872857 | 690 | + | PROTEIN:amino acid transporters |
| LCRIS_00871 | LCRIS_00871 | 872884 | 873057 | 174 | + | PROTEIN:hypothetical protein |
| LCRIS_00872 | dapF | 873101 | 874105 | 1005 | - | PROTEIN:diaminopimelate epimerase |
| LCRIS_00873 | lysC1 | 874114 | 875469 | 1356 | - | PROTEIN:aspartokinase |
| LCRIS_00874 | lysA | 875928 | 877238 | 1311 | + | PROTEIN:diaminopimelate decarboxylase |
| LCRIS_00875 | dapD | 877253 | 877960 | 708 | + | PROTEIN:2,3,4,5-tetrahydropyridine-2-carboxylate n |
| LCRIS_00876 | LCRIS_00876 | 877962 | 879116 | 1155 | + | PROTEIN:peptidase, m20/m25/m40 family |
| LCRIS_00877 | dapA | 879118 | 880044 | 927 | + | PROTEIN:dihydrodipicolinate synthase |
| LCRIS_00878 | dapB | 880051 | 880830 | 780 | + | PROTEIN:dihydrodipicolinate reductase |
| LCRIS_00879 | patA1 | 880842 | 882017 | 1176 | + | PROTEIN:aspartate aminotransferase |
| LCRIS_00880 | asd | 882017 | 883075 | 1059 | + | PROTEIN:aspartate-semialdehyde dehydrogenase |
| LCRIS_00881 | LCRIS_00881 | 883238 | 884281 | 1044 | + | PROTEIN:penicillin-binding protein |
| LCRIS_00882 | LCRIS_00882 | 884324 | 885934 | 1611 | - | PROTEIN:triacylglycerol lipase |
| LCRIS_00883 | LCRIS_00883 | 886270 | 886536 | 267 | + | PROTEIN:polyferredoxin |
| LCRIS_00884 | LCRIS_00884 | 886546 | 887061 | 516 | + | PROTEIN:hypothetical protein |
| LCRIS_00885 | iunH2 | 887087 | 888013 | 927 | - | PROTEIN:inosine-uridine preferring nucleoside hydr |
| LCRIS_00886 | LCRIS_00886 | 888144 | 888416 | 273 | + | PROTEIN:hypothetical protein |
| LCRIS_00887 | LCRIS_00887 | 888532 | 888801 | 270 | + | PROTEIN:DNA alkylation repair enzyme |
| LCRIS_00888 | bglA2 | 888866 | 890314 | 1449 | - | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_00889 | LCRIS_00889 | 890401 | 891105 | 705 | - | PROTEIN:transcriptional regulator |
| LCRIS_00890 | celA1 | 891471 | 891809 | 339 | + | PROTEIN:PTS system cellobiose-specific iib compone |
| LCRIS_00891 | celC1 | 891850 | 892221 | 372 | + | PROTEIN:PTS system cellobiose-specific iia compone |
| LCRIS_00892 | LCRIS_00892 | 892317 | 892844 | 528 | + | PROTEIN:hypothetical protein |
| LCRIS_00893 | celB3 | 893017 | 894369 | 1353 | + | PROTEIN:PTS system cellobiose-specific iic compone |
| LCRIS_00894 | LCRIS_00894 | 894477 | 894887 | 411 | - | PROTEIN:hypothetical protein |
| LCRIS_00895 | bglA3 | 894990 | 896471 | 1482 | - | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_00896 | ydhQ | 896481 | 897203 | 723 | - | PROTEIN:transcriptional regulator, gntr family |
| LCRIS_00897 | LCRIS_00897 | 897325 | 897567 | 243 | - | PROTEIN:hypothetical protein |
| LCRIS_00898 | celB4 | 897586 | 899001 | 1416 | - | PROTEIN:PTS system cellobiose-specific iic compone |
| LCRIS_00899 | bglA4 | 899001 | 900383 | 1383 | - | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_00900 | glk | 900554 | 901414 | 861 | - | PROTEIN:sugar kinase, rok family |
| LCRIS_00901 | LCRIS_00901 | 901533 | 902021 | 489 | + | PROTEIN:nitroreductase |
| LCRIS_00902 | LCRIS_00902 | 902128 | 902775 | 648 | + | PROTEIN:hypothetical protein |
| LCRIS_00903 | eno | 902855 | 904141 | 1287 | - | PROTEIN:enolase |
| LCRIS_00904 | LCRIS_00904 | 904356 | 905189 | 834 | + | PROTEIN:degv family protein |
| LCRIS_00905 | LCRIS_00905 | 905222 | 905797 | 576 | - | PROTEIN:phospholipid phosphatase |
| LCRIS_00906 | cbh | 905901 | 906878 | 978 | - | PROTEIN:choloylglycine hydrolase |
| LCRIS_02096 | tRNA-Ser | 907005 | 907093 | 89 | - | RNA:Ser tRNA |
| LCRIS_00907 | LCRIS_00907 | 907274 | 908305 | 1032 | + | PROTEIN:hypothetical protein |
| LCRIS_00908 | dps | 908433 | 908900 | 468 | + | PROTEIN:non-heme iron-containing ferritin |
| LCRIS_00909 | LCRIS_00909 | 908993 | 909205 | 213 | + | PROTEIN:hypothetical protein |
| LCRIS_00910 | LCRIS_00910 | 909171 | 909899 | 729 | + | PROTEIN:succinate dehydrogenase/fumarate reductase |
| LCRIS_00911 | LCRIS_00911 | 909913 | 910152 | 240 | + | PROTEIN:hypothetical protein |
| LCRIS_00912 | LCRIS_00912 | 910442 | 911038 | 597 | + | PROTEIN:had superfamily hydrolase |
| LCRIS_00913 | LCRIS_00913 | 911198 | 911944 | 747 | + | PROTEIN:hypothetical protein |
| LCRIS_00914 | yjaB | 912714 | 913163 | 450 | - | PROTEIN:acetyltransferase, gnat family |
| LCRIS_00915 | LCRIS_00915 | 913391 | 914740 | 1350 | + | PROTEIN:major facilitator superfamily, na-driven e |
| LCRIS_00916 | LCRIS_00916 | 915084 | 915572 | 489 | + | PROTEIN:transposase |
| LCRIS_00917 | LCRIS_00917 | 915562 | 915717 | 156 | + | PROTEIN:hypothetical protein |
| LCRIS_00918 | LCRIS_00918 | 916049 | 916450 | 402 | + | PROTEIN:hypothetical protein |
| LCRIS_00919 | LCRIS_00919 | 916465 | 925272 | 8808 | + | PROTEIN:mucus-binding protein |
| LCRIS_00920 | LCRIS_00920 | 925505 | 926911 | 1407 | + | PROTEIN:transposase |
| LCRIS_00921 | LCRIS_00921 | 927057 | 927908 | 852 | + | PROTEIN:hypothetical protein |
| LCRIS_00922 | LCRIS_00922 | 928049 | 929356 | 1308 | + | PROTEIN:glycosyl transferase, family 2 |
| LCRIS_00923 | LCRIS_00923 | 929356 | 930453 | 1098 | + | PROTEIN:endoglucanase |
| LCRIS_00924 | LCRIS_00924 | 930820 | 931095 | 276 | + | PROTEIN:transposase |
| LCRIS_00925 | LCRIS_00925 | 931120 | 931275 | 156 | + | PROTEIN:hypothetical protein |
| LCRIS_00926 | LCRIS_00926 | 931845 | 932642 | 798 | + | PROTEIN:RNA-directed DNA polymerase |
| LCRIS_00927 | ribH | 932720 | 933184 | 465 | - | PROTEIN:riboflavin synthase, beta subunit |
| LCRIS_00928 | ribA | 933199 | 934377 | 1179 | - | PROTEIN:GTP cyclohydrolase ii |
| LCRIS_00929 | ribE | 934379 | 934975 | 597 | - | PROTEIN:riboflavin synthase, alpha subunit |
| LCRIS_00930 | ribD | 934968 | 936026 | 1059 | - | PROTEIN:riboflavin biosynthesis protein ribd |
| LCRIS_00931 | LCRIS_00931 | 936113 | 936301 | 189 | + | PROTEIN:hypothetical protein |
| LCRIS_00932 | LCRIS_00932 | 936453 | 937247 | 795 | + | PROTEIN:short-chain dehydrogenase/reductase sdr |
| LCRIS_00933 | LCRIS_00933 | 937311 | 937664 | 354 | - | PROTEIN:hypothetical protein |
| LCRIS_00934 | LCRIS_00934 | 937726 | 938163 | 438 | - | PROTEIN:response regulator of the lytr/algr family |
| LCRIS_00935 | LCRIS_00935 | 938322 | 939185 | 864 | - | PROTEIN:transcriptional regulator |
| LCRIS_00936 | LCRIS_00936 | 939313 | 939891 | 579 | + | PROTEIN:alpha/beta hydrolase superfamily protein |
| LCRIS_00937 | LCRIS_00937 | 940151 | 941662 | 1512 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_00938 | LCRIS_00938 | 941965 | 942648 | 684 | - | PROTEIN:integral hypothetical protein |
| LCRIS_00939 | LCRIS_00939 | 942809 | 943270 | 462 | + | PROTEIN:hypothetical protein |
| LCRIS_00940 | LCRIS_00940 | 943257 | 943880 | 624 | + | PROTEIN:mutt/nudix family protein |
| LCRIS_00941 | brnQ | 944266 | 945642 | 1377 | + | PROTEIN:branched-chain amino acid transport protei |
| LCRIS_00942 | LCRIS_00942 | 945759 | 947498 | 1740 | + | PROTEIN:hypothetical protein |
| LCRIS_00943 | tpx | 947551 | 948045 | 495 | - | PROTEIN:thiol peroxidase |
| LCRIS_00944 | LCRIS_00944 | 948224 | 948382 | 159 | + | PROTEIN:hypothetical protein |
| LCRIS_00945 | LCRIS_00945 | 948480 | 948716 | 237 | - | PROTEIN:hypothetical protein |
| LCRIS_00946 | LCRIS_00946 | 948757 | 949506 | 750 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_00947 | LCRIS_00947 | 949600 | 950682 | 1083 | + | PROTEIN:oxidoreductase |
| LCRIS_00948 | LCRIS_00948 | 950772 | 951263 | 492 | + | PROTEIN:acetyltransferase, gnat family |
| LCRIS_00949 | LCRIS_00949 | 951265 | 951921 | 657 | + | PROTEIN:glucose-6-phosphate 1-dehydrogenase |
| LCRIS_00950 | LCRIS_00950 | 952361 | 952600 | 240 | + | PROTEIN:hypothetical protein |
| LCRIS_00951 | LCRIS_00951 | 952822 | 954417 | 1596 | + | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_00952 | LCRIS_00952 | 954414 | 955997 | 1584 | + | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_00953 | LCRIS_00953 | 956127 | 957518 | 1392 | + | PROTEIN:amino acid permease |
| LCRIS_00954 | thyA | 957680 | 958591 | 912 | + | PROTEIN:thymidylate synthase |
| LCRIS_00955 | folA | 958601 | 959107 | 507 | + | PROTEIN:dihydrofolate reductase |
| LCRIS_00956 | LCRIS_00956 | 959197 | 961599 | 2403 | + | PROTEIN:cation-transporting atpase |
| LCRIS_00957 | LCRIS_00957 | 961641 | 962582 | 942 | - | PROTEIN:DNA-entry nuclease |
| LCRIS_00958 | metQ1 | 963001 | 963855 | 855 | + | PROTEIN:ABC transporter, substrate binding protein |
| LCRIS_00959 | metN | 963868 | 964932 | 1065 | + | PROTEIN:methionine import ATP-binding protein metn |
| LCRIS_00960 | metI | 964925 | 965644 | 720 | + | PROTEIN:ABC transporter, permease protein |
| LCRIS_00961 | fumC | 965775 | 967178 | 1404 | + | PROTEIN:fumarate hydratase |
| LCRIS_00962 | frdA2 | 967191 | 968567 | 1377 | + | PROTEIN:fumarate reductase, flavoprotein subunit |
| LCRIS_00963 | ldh2 | 968764 | 969693 | 930 | + | PROTEIN:L-lactate dehydrogenase |
| LCRIS_00964 | pepC4 | 969831 | 971147 | 1317 | + | PROTEIN:aminopeptidase c |
| LCRIS_00965 | hxlA | 971399 | 972097 | 699 | + | PROTEIN:dimethylmenaquinone methyltransferase |
| LCRIS_00966 | LCRIS_00966 | 972114 | 973583 | 1470 | + | PROTEIN:2-oxoglutarate/malate translocator |
| LCRIS_00967 | LCRIS_00967 | 973665 | 974456 | 792 | + | PROTEIN:hydrolase or acyltransferase |
| LCRIS_00968 | citC | 974537 | 975592 | 1056 | + | PROTEIN:citrate lyase ligase |
| LCRIS_00969 | citD | 975582 | 975875 | 294 | + | PROTEIN:citrate lyase acyl carrier protein |
| LCRIS_00970 | citE | 975876 | 976790 | 915 | + | PROTEIN:citrate lyase, beta subunit |
| LCRIS_00971 | citF | 976780 | 978321 | 1542 | + | PROTEIN:citrate lyase, alpha subunit |
| LCRIS_00972 | LCRIS_00972 | 978657 | 979112 | 456 | + | PROTEIN:hypothetical protein |
| LCRIS_00973 | LCRIS_00973 | 979142 | 979303 | 162 | + | PROTEIN:hypothetical protein |
| LCRIS_00974 | cbiQ3 | 979321 | 980154 | 834 | - | PROTEIN:cobalt ABC transporter permease |
| LCRIS_00975 | cbiO4 | 980147 | 981856 | 1710 | - | PROTEIN:cobalt ABC transporter ATP-binding protein |
| LCRIS_00976 | LCRIS_00976 | 981860 | 982417 | 558 | - | PROTEIN:hypothetical protein |
| LCRIS_00977 | LCRIS_00977 | 982575 | 982955 | 381 | + | PROTEIN:hypothetical protein |
| LCRIS_00978 | LCRIS_00978 | 983256 | 983582 | 327 | - | PROTEIN:transporter protein |
| LCRIS_00979 | citR | 983630 | 984568 | 939 | - | PROTEIN:citrate lyase regulator |
| LCRIS_00980 | LCRIS_00980 | 984732 | 986078 | 1347 | + | PROTEIN:hypothetical protein |
| LCRIS_00981 | ebsC | 986478 | 986987 | 510 | - | PROTEIN:transcriptional regulator |
| LCRIS_00982 | prmA | 987048 | 987992 | 945 | + | PROTEIN:ribosomal protein l11 methyltransferase |
| LCRIS_00983 | LCRIS_00983 | 988048 | 988410 | 363 | + | PROTEIN:hypothetical protein |
| LCRIS_00984 | relA | 988425 | 990665 | 2241 | + | PROTEIN:GTP pyrophosphokinase |
| LCRIS_00985 | dtd | 990665 | 991102 | 438 | + | PROTEIN:d-tyrosyl-tRNA(tyr) deacylase |
| LCRIS_00986 | hisS | 991398 | 992684 | 1287 | + | PROTEIN:histidyl-tRNA synthetase |
| LCRIS_00987 | aspS | 992690 | 994543 | 1854 | + | PROTEIN:aspartyl-tRNA synthetase |
| LCRIS_00988 | patA2 | 994610 | 995794 | 1185 | - | PROTEIN:aspartate aminotransferase |
| LCRIS_00989 | LCRIS_00989 | 995913 | 996170 | 258 | - | PROTEIN:hypothetical protein |
| LCRIS_00990 | coaBC | 996272 | 997462 | 1191 | + | PROTEIN:phosphopantothenoylcysteine decarboxylase/ |
| LCRIS_00991 | LCRIS_00991 | 997977 | 998813 | 837 | + | PROTEIN:oxidoreductase, aldo/keto reductase family |
| LCRIS_00992 | LCRIS_00992 | 998862 | 999674 | 813 | + | PROTEIN:cof-like hydrolase |
| LCRIS_00993 | LCRIS_00993 | 999741 | 1000706 | 966 | + | PROTEIN:d-isomer specific 2-hydroxyacid dehydrogen |
| LCRIS_00994 | LCRIS_00994 | 1000773 | 1002158 | 1386 | - | PROTEIN:amino acid permease |
| LCRIS_00995 | LCRIS_00995 | 1002256 | 1002870 | 615 | - | PROTEIN:alkaline phosphatase |
| LCRIS_00996 | uvrC | 1003010 | 1004812 | 1803 | + | PROTEIN:uvrABC system protein c |
| LCRIS_00997 | obg | 1004891 | 1006195 | 1305 | + | PROTEIN:GTP-binding protein |
| LCRIS_00998 | LCRIS_00998 | 1006211 | 1007503 | 1293 | + | PROTEIN:acyltransferase family protein |
| LCRIS_00999 | LCRIS_00999 | 1007680 | 1008126 | 447 | + | PROTEIN:esterase |
| LCRIS_01000 | elaC | 1008144 | 1009082 | 939 | + | PROTEIN:ribonuclease z |
| LCRIS_01001 | LCRIS_01001 | 1009079 | 1009873 | 795 | + | PROTEIN:oxidoreductase, short-chain dehydrogenase/ |
| LCRIS_01002 | LCRIS_01002 | 1009880 | 1010128 | 249 | + | PROTEIN:hypothetical protein |
| LCRIS_01003 | rpmF | 1010231 | 1010419 | 189 | + | PROTEIN:50S ribosomal protein l32 |
| LCRIS_01004 | LCRIS_01004 | 1010479 | 1012035 | 1557 | + | PROTEIN:2',3'-cyclic-nucleotide 2'-phosphodiestera |
| LCRIS_01005 | LCRIS_01005 | 1012097 | 1012273 | 177 | - | PROTEIN:hypothetical protein |
| LCRIS_01006 | dnaE | 1012431 | 1015538 | 3108 | + | PROTEIN:DNA polymerase iii, alpha subunit |
| LCRIS_01007 | pfk | 1015706 | 1016668 | 963 | + | PROTEIN:6-phosphofructokinase |
| LCRIS_01008 | pyk | 1016702 | 1018471 | 1770 | + | PROTEIN:pyruvate kinase |
| LCRIS_01009 | LCRIS_01009 | 1018598 | 1019491 | 894 | + | PROTEIN:RNA-binding protein, s1-like domain |
| LCRIS_01010 | xerD | 1019478 | 1020383 | 906 | + | PROTEIN:tyrosine recombinase xerd |
| LCRIS_01011 | ribT | 1020392 | 1020754 | 363 | + | PROTEIN:reductase |
| LCRIS_01012 | scpA | 1020741 | 1021481 | 741 | + | PROTEIN:segregation and condensation protein a |
| LCRIS_01013 | scpB | 1021471 | 1022061 | 591 | + | PROTEIN:segregation and condensation protein b |
| LCRIS_01014 | rluB | 1022065 | 1022781 | 717 | + | PROTEIN:pseudouridine synthase |
| LCRIS_01015 | LCRIS_01015 | 1022999 | 1023688 | 690 | + | PROTEIN:hypothetical protein |
| LCRIS_01016 | LCRIS_01016 | 1023790 | 1024251 | 462 | + | PROTEIN:N-acetylmuramidase |
| LCRIS_01017 | cmk | 1024303 | 1024974 | 672 | + | PROTEIN:cytidylate kinase |
| LCRIS_01018 | rpsA | 1025047 | 1026255 | 1209 | + | PROTEIN:30S ribosomal protein s1 |
| LCRIS_01019 | engA | 1026325 | 1027632 | 1308 | + | PROTEIN:GTP-binding protein enga |
| LCRIS_01020 | hupB1 | 1027791 | 1028066 | 276 | + | PROTEIN:DNA-binding protein hu |
| LCRIS_01021 | LCRIS_01021 | 1028147 | 1029394 | 1248 | + | PROTEIN:o-linked transferase |
| LCRIS_01022 | cls2 | 1029435 | 1030925 | 1491 | - | PROTEIN:cardiolipin synthetase |
| LCRIS_01023 | LCRIS_01023 | 1031086 | 1032666 | 1581 | - | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_01024 | LCRIS_01024 | 1032805 | 1033140 | 336 | + | PROTEIN:hypothetical protein |
| LCRIS_01025 | LCRIS_01025 | 1033193 | 1033732 | 540 | - | PROTEIN:acetyltransferase, gnat family |
| LCRIS_01026 | LCRIS_01026 | 1033736 | 1034194 | 459 | - | PROTEIN:hypothetical protein |
| LCRIS_01027 | LCRIS_01027 | 1034352 | 1035161 | 810 | + | PROTEIN:hypothetical protein |
| LCRIS_01028 | LCRIS_01028 | 1035173 | 1035475 | 303 | + | PROTEIN:pyrophosphatase |
| LCRIS_01029 | LCRIS_01029 | 1035468 | 1035833 | 366 | + | PROTEIN:hypothetical protein |
| LCRIS_01030 | glsA | 1035960 | 1036733 | 774 | + | PROTEIN:glutaminase |
| LCRIS_01031 | LCRIS_01031 | 1036833 | 1038215 | 1383 | + | PROTEIN:fibronectin-binding protein |
| LCRIS_01032 | LCRIS_01032 | 1038472 | 1038993 | 522 | + | PROTEIN:acetyltransferase |
| LCRIS_01033 | LCRIS_01033 | 1039033 | 1039815 | 783 | + | PROTEIN:hydrolases or acyltransferases |
| LCRIS_01034 | LCRIS_01034 | 1040145 | 1040396 | 252 | + | PROTEIN:hypothetical protein |
| LCRIS_01035 | LCRIS_01035 | 1040615 | 1041031 | 417 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01036 | cofE | 1041125 | 1041838 | 714 | + | PROTEIN:hypothetical protein |
| LCRIS_01037 | LCRIS_01037 | 1041864 | 1042301 | 438 | + | PROTEIN:uridine kinase |
| LCRIS_01038 | LCRIS_01038 | 1042404 | 1043165 | 762 | + | PROTEIN:phosphorylase, pnp/udp family |
| LCRIS_01039 | LCRIS_01039 | 1043197 | 1043961 | 765 | + | PROTEIN:phosphorylase, pnp/udp family |
| LCRIS_01040 | LCRIS_01040 | 1044016 | 1044819 | 804 | - | PROTEIN:aminoglycoside n3'-acetyltransferase |
| LCRIS_01041 | LCRIS_01041 | 1045105 | 1045596 | 492 | - | PROTEIN:amino acid permease-associated region |
| LCRIS_01042 | LCRIS_01042 | 1045621 | 1046127 | 507 | - | PROTEIN:amino acid transporters |
| LCRIS_01043 | LCRIS_01043 | 1046158 | 1046415 | 258 | - | PROTEIN:amino acid transporters |
| LCRIS_01044 | LCRIS_01044 | 1046641 | 1047141 | 501 | + | PROTEIN:lactocepin s-layer protein |
| LCRIS_01045 | LCRIS_01045 | 1047185 | 1048069 | 885 | - | PROTEIN:hypothetical protein |
| LCRIS_01046 | cca | 1048158 | 1049357 | 1200 | + | PROTEIN:cca-adding enzyme |
| LCRIS_01047 | hlyIII | 1049394 | 1050059 | 666 | - | PROTEIN:hemolysin iii |
| LCRIS_01048 | LCRIS_01048 | 1050173 | 1051024 | 852 | + | PROTEIN:degv family protein |
| LCRIS_01049 | LCRIS_01049 | 1051027 | 1051257 | 231 | + | PROTEIN:hypothetical protein |
| LCRIS_01050 | LCRIS_01050 | 1051306 | 1051731 | 426 | + | PROTEIN:hypothetical protein |
| LCRIS_01051 | LCRIS_01051 | 1051676 | 1052722 | 1047 | + | PROTEIN:Na+/H+ antiporter |
| LCRIS_01052 | LCRIS_01052 | 1052792 | 1053631 | 840 | + | PROTEIN:GTP-binding protein |
| LCRIS_01053 | rnhB | 1053628 | 1054380 | 753 | + | PROTEIN:ribonuclease hii |
| LCRIS_01054 | smf | 1054442 | 1055290 | 849 | + | PROTEIN:DNA processing protein |
| LCRIS_01055 | topA | 1055360 | 1057471 | 2112 | + | PROTEIN:DNA topoisomerase |
| LCRIS_01056 | trmFO | 1057494 | 1058810 | 1317 | + | PROTEIN:methylenetetrahydrofolate--tRNA-(uracil-5- |
| LCRIS_01057 | xerC | 1058810 | 1059421 | 612 | + | PROTEIN:integrase/recombinase |
| LCRIS_01058 | LCRIS_01058 | 1059381 | 1059719 | 339 | + | PROTEIN:integrase/recombinase |
| LCRIS_01059 | hslV | 1059728 | 1060252 | 525 | + | PROTEIN:ATP-dependent protease hslvu, proteolytic |
| LCRIS_01060 | hslU | 1060264 | 1061661 | 1398 | + | PROTEIN:ATP-dependent protease hslvu, atpase subun |
| LCRIS_01061 | LCRIS_01061 | 1061821 | 1062717 | 897 | + | PROTEIN:galactose mutarotase related enzyme |
| LCRIS_01062 | LCRIS_01062 | 1062778 | 1063200 | 423 | + | PROTEIN:aldo/keto reductase |
| LCRIS_01063 | LCRIS_01063 | 1063197 | 1063460 | 264 | + | PROTEIN:2,5-diketo-d-gluconic acid reductase a |
| LCRIS_01064 | LCRIS_01064 | 1063748 | 1064440 | 693 | + | PROTEIN:aggregation promoting factor |
| LCRIS_01065 | LCRIS_01065 | 1064537 | 1065352 | 816 | - | PROTEIN:integrase-recombinase |
| LCRIS_01066 | crcB1 | 1065617 | 1066015 | 399 | + | PROTEIN:protein crcb homolog |
| LCRIS_01067 | crcB2 | 1066012 | 1066389 | 378 | + | PROTEIN:protein crcb homolog |
| LCRIS_01068 | LCRIS_01068 | 1066409 | 1066630 | 222 | + | PROTEIN:homocysteine s-methyltransferase |
| LCRIS_01069 | LCRIS_01069 | 1066624 | 1067367 | 744 | + | PROTEIN:amino acid permease |
| LCRIS_01070 | pepV | 1067503 | 1068909 | 1407 | - | PROTEIN:xaa-his dipeptidase |
| LCRIS_01071 | LCRIS_01071 | 1069096 | 1069602 | 507 | + | PROTEIN:hypothetical protein |
| LCRIS_01072 | LCRIS_01072 | 1069650 | 1071122 | 1473 | - | PROTEIN:amino acid transporter |
| LCRIS_01073 | speC | 1071355 | 1073442 | 2088 | + | PROTEIN:ornithine decarboxylase |
| LCRIS_01074 | LCRIS_01074 | 1073453 | 1074712 | 1260 | + | PROTEIN:aluminum resistance protein |
| LCRIS_01075 | recQ | 1074832 | 1076601 | 1770 | + | PROTEIN:ATP-dependent DNA helicase recq |
| LCRIS_01076 | LCRIS_01076 | 1076682 | 1077134 | 453 | + | PROTEIN:trp repressor binding protein |
| LCRIS_01077 | uvrA2 | 1077545 | 1080058 | 2514 | + | PROTEIN:uvrABC system protein a |
| LCRIS_01078 | mmuM | 1080220 | 1081209 | 990 | + | PROTEIN:homocysteine s-methyltransferase |
| LCRIS_01079 | LCRIS_01079 | 1081206 | 1081433 | 228 | + | PROTEIN:hypothetical protein |
| LCRIS_01080 | LCRIS_01080 | 1081465 | 1082097 | 633 | - | PROTEIN:phospholipid phosphatase |
| LCRIS_01081 | LCRIS_01081 | 1082196 | 1082645 | 450 | + | PROTEIN:acetyltransferase, gnat family |
| LCRIS_01082 | maa3 | 1082662 | 1083276 | 615 | + | PROTEIN:maltose o-acetyltransferase |
| LCRIS_01083 | pbpX | 1083458 | 1084528 | 1071 | + | PROTEIN:penicillin-binding protein |
| LCRIS_01084 | LCRIS_01084 | 1084576 | 1084875 | 300 | + | PROTEIN:hypothetical protein |
| LCRIS_01085 | LCRIS_01085 | 1084836 | 1085135 | 300 | - | PROTEIN:hypothetical protein |
| LCRIS_01086 | LCRIS_01086 | 1085183 | 1085362 | 180 | - | PROTEIN:hypothetical protein |
| LCRIS_01087 | LCRIS_01087 | 1085398 | 1085598 | 201 | + | PROTEIN:hypothetical protein |
| LCRIS_01088 | pdxK | 1085646 | 1086263 | 618 | + | PROTEIN:pyridoxal kinase |
| LCRIS_01089 | LCRIS_01089 | 1086316 | 1086495 | 180 | + | PROTEIN:hypothetical protein |
| LCRIS_01090 | LCRIS_01090 | 1086498 | 1087064 | 567 | + | PROTEIN:hypothetical protein |
| LCRIS_01091 | LCRIS_01091 | 1087139 | 1088344 | 1206 | + | PROTEIN:secreted protein |
| LCRIS_01092 | LCRIS_01092 | 1088790 | 1089245 | 456 | + | PROTEIN:acetyltransferase, gnat family |
| LCRIS_01093 | pepT1 | 1089258 | 1090541 | 1284 | + | PROTEIN:peptidase t |
| LCRIS_01094 | LCRIS_01094 | 1090895 | 1091848 | 954 | - | PROTEIN:atpase, aaa family |
| LCRIS_01095 | LCRIS_01095 | 1091862 | 1092158 | 297 | - | PROTEIN:atpase, aaa family |
| LCRIS_01096 | spxA | 1092300 | 1092701 | 402 | + | PROTEIN:regulatory protein spx |
| LCRIS_01097 | treC | 1092919 | 1094271 | 1353 | + | PROTEIN:alpha,alpha-phosphotrehalase |
| LCRIS_01098 | LCRIS_01098 | 1094449 | 1094886 | 438 | + | PROTEIN:hypothetical protein |
| LCRIS_01099 | ugpB | 1095221 | 1096498 | 1278 | - | PROTEIN:glycerol-3-phosphate ABC transporter |
| LCRIS_01100 | LCRIS_01100 | 1096860 | 1097225 | 366 | + | PROTEIN:pyridoxine 5'-phosphate oxidase v related |
| LCRIS_01101 | LCRIS_01101 | 1097329 | 1098555 | 1227 | + | PROTEIN:phosphohydrolase |
| LCRIS_01102 | LCRIS_01102 | 1099121 | 1099279 | 159 | + | PROTEIN:lpxtg-motif cell wall anchor |
| LCRIS_01103 | LCRIS_01103 | 1099424 | 1101517 | 2094 | + | PROTEIN:hypothetical protein |
| LCRIS_01104 | hsdR | 1101656 | 1104874 | 3219 | + | PROTEIN:type i restriction-modification system, r |
| LCRIS_01105 | LCRIS_01105 | 1104970 | 1106172 | 1203 | + | PROTEIN:abortive phage resistance protein |
| LCRIS_01106 | hsdM | 1106242 | 1107699 | 1458 | + | PROTEIN:type i restriction-modification enzyme, m |
| LCRIS_01107 | hsdS1 | 1107689 | 1108405 | 717 | + | PROTEIN:type i site-specific deoxyribonuclease |
| LCRIS_01108 | hsdS2 | 1108416 | 1109108 | 693 | + | PROTEIN:type i restriction-modification system, s |
| LCRIS_01109 | LCRIS_01109 | 1109240 | 1109863 | 624 | + | PROTEIN:hypothetical protein |
| LCRIS_01110 | LCRIS_01110 | 1109874 | 1110302 | 429 | + | PROTEIN:hypothetical protein |
| LCRIS_01111 | LCRIS_01111 | 1110336 | 1111187 | 852 | + | PROTEIN:hypothetical protein |
| LCRIS_01112 | LCRIS_01112 | 1111269 | 1112189 | 921 | - | PROTEIN:hypothetical protein |
| LCRIS_01113 | LCRIS_01113 | 1112319 | 1114106 | 1788 | + | PROTEIN:hypothetical protein |
| LCRIS_01114 | LCRIS_01114 | 1114167 | 1114337 | 171 | + | PROTEIN:hypothetical protein |
| LCRIS_01115 | LCRIS_01115 | 1114413 | 1114691 | 279 | + | PROTEIN:hypothetical protein |
| LCRIS_01116 | LCRIS_01116 | 1114845 | 1115405 | 561 | + | PROTEIN:hypothetical protein |
| LCRIS_01117 | LCRIS_01117 | 1115539 | 1116111 | 573 | + | PROTEIN:tpr repeat protein |
| LCRIS_01118 | LCRIS_01118 | 1116187 | 1116633 | 447 | + | PROTEIN:hypothetical protein |
| LCRIS_01119 | LCRIS_01119 | 1116669 | 1116881 | 213 | - | PROTEIN:hypothetical protein |
| LCRIS_01120 | LCRIS_01120 | 1117056 | 1118084 | 1029 | + | PROTEIN:hypothetical protein |
| LCRIS_01121 | LCRIS_01121 | 1118086 | 1118658 | 573 | + | PROTEIN:hypothetical protein |
| LCRIS_01122 | LCRIS_01122 | 1118699 | 1119277 | 579 | + | PROTEIN:tpr repeat protein |
| LCRIS_01123 | LCRIS_01123 | 1119618 | 1120532 | 915 | + | PROTEIN:mucus-binding protein |
| LCRIS_01124 | psd | 1120540 | 1121175 | 636 | + | PROTEIN:phosphatidylserine decarboxylase |
| LCRIS_01125 | LCRIS_01125 | 1121312 | 1121647 | 336 | + | PROTEIN:hypothetical protein |
| LCRIS_01126 | LCRIS_01126 | 1121647 | 1121808 | 162 | + | PROTEIN:formyltetrahydrofolate synthetase |
| LCRIS_01127 | fhs | 1121880 | 1123322 | 1443 | + | PROTEIN:formate--tetrahydrofolate ligase |
| LCRIS_01128 | lspA | 1123328 | 1123792 | 465 | + | PROTEIN:lipoprotein signal peptidase |
| LCRIS_01129 | rluD2 | 1123785 | 1124696 | 912 | + | PROTEIN:pseudouridine synthase |
| LCRIS_01130 | carA1 | 1124701 | 1125756 | 1056 | + | PROTEIN:carbamoyl-phosphate synthase small subunit |
| LCRIS_01131 | carB1 | 1125761 | 1128955 | 3195 | + | PROTEIN:carbamoyl-phosphate synthase large subunit |
| LCRIS_01132 | LCRIS_01132 | 1129030 | 1130721 | 1692 | - | PROTEIN:fibronectin-binding protein |
| LCRIS_01133 | LCRIS_01133 | 1131004 | 1131420 | 417 | + | PROTEIN:l-fucose operon regulator |
| LCRIS_01134 | degV2 | 1131490 | 1132368 | 879 | + | PROTEIN:degv family protein |
| LCRIS_01135 | LCRIS_01135 | 1132704 | 1134419 | 1716 | + | PROTEIN:muts family protein |
| LCRIS_01136 | LCRIS_01136 | 1134401 | 1134580 | 180 | + | PROTEIN:hypothetical protein |
| LCRIS_01137 | xerC | 1134634 | 1135701 | 1068 | - | PROTEIN:tyrosine recombinase xerc |
| LCRIS_01138 | LCRIS_01138 | 1136204 | 1136662 | 459 | + | PROTEIN:hypothetical protein |
| LCRIS_01139 | LCRIS_01139 | 1136707 | 1137318 | 612 | - | PROTEIN:diaphanous protein |
| LCRIS_01140 | LCRIS_01140 | 1137468 | 1138625 | 1158 | - | PROTEIN:lysin |
| LCRIS_01141 | LCRIS_01141 | 1138983 | 1139243 | 261 | + | PROTEIN:transglycosylase-associated protein |
| LCRIS_01142 | LCRIS_01142 | 1140612 | 1141307 | 696 | - | PROTEIN:hypothetical protein |
| LCRIS_01143 | LCRIS_01143 | 1141615 | 1141974 | 360 | + | PROTEIN:transcriptional regulator, xre family |
| LCRIS_01144 | ppiB | 1142016 | 1142615 | 600 | - | PROTEIN:peptidyl-prolyl cis-trans isomerase |
| LCRIS_01145 | ppaC | 1142701 | 1143636 | 936 | - | PROTEIN:manganese-dependent inorganic pyrophosphat |
| LCRIS_01146 | LCRIS_01146 | 1143666 | 1144640 | 975 | - | PROTEIN:transcriptional regulator |
| LCRIS_01147 | parC | 1144788 | 1147244 | 2457 | - | PROTEIN:DNA topoisomerase iv, a subunit |
| LCRIS_01148 | parE | 1147263 | 1149209 | 1947 | - | PROTEIN:DNA topoisomerase iv, b subunit |
| LCRIS_01149 | ygiH | 1149308 | 1149943 | 636 | + | PROTEIN:hypothetical protein |
| LCRIS_01150 | LCRIS_01150 | 1150061 | 1153117 | 3057 | - | PROTEIN:non-specific serine/threonine protein kina |
| LCRIS_01151 | LCRIS_01151 | 1153554 | 1154732 | 1179 | + | PROTEIN:transposase |
| LCRIS_01152 | LCRIS_01152 | 1154894 | 1155118 | 225 | - | PROTEIN:replication initiator a, n-terminal |
| LCRIS_01153 | LCRIS_01153 | 1155292 | 1155843 | 552 | - | PROTEIN:isochorismatase family protein |
| LCRIS_01154 | LCRIS_01154 | 1155855 | 1156610 | 756 | - | PROTEIN:integral hypothetical protein |
| LCRIS_01155 | LCRIS_01155 | 1156657 | 1157313 | 657 | - | PROTEIN:hypothetical protein |
| LCRIS_01156 | LCRIS_01156 | 1157339 | 1157947 | 609 | - | PROTEIN:hypothetical protein |
| LCRIS_01157 | LCRIS_01157 | 1158045 | 1158401 | 357 | - | PROTEIN:hypothetical protein |
| LCRIS_01158 | LCRIS_01158 | 1158531 | 1158764 | 234 | + | PROTEIN:hypothetical protein |
| LCRIS_01159 | LCRIS_01159 | 1159185 | 1159808 | 624 | + | PROTEIN:nucleoside-triphosphatase |
| LCRIS_01160 | LCRIS_01160 | 1159939 | 1160376 | 438 | - | PROTEIN:hypothetical protein |
| LCRIS_01161 | LCRIS_01161 | 1160426 | 1161658 | 1233 | - | PROTEIN:3',5'-cyclic-nucleotide phosphodiesterase |
| LCRIS_01162 | ycbY | 1161662 | 1162786 | 1125 | - | PROTEIN:n6-adenine-specific DNA methylase |
| LCRIS_01163 | LCRIS_01163 | 1163247 | 1163657 | 411 | - | PROTEIN:cell division initiation protein |
| LCRIS_01164 | LCRIS_01164 | 1163751 | 1164320 | 570 | - | PROTEIN:hypothetical protein |
| LCRIS_01165 | recU | 1164389 | 1165018 | 630 | + | PROTEIN:holliday junction resolvase recu |
| LCRIS_01166 | mrcA | 1165021 | 1167333 | 2313 | + | PROTEIN:penicillin-binding protein 1a |
| LCRIS_01167 | nth | 1167488 | 1168117 | 630 | - | PROTEIN:endonuclease iii |
| LCRIS_01168 | dnaD | 1168120 | 1168764 | 645 | - | PROTEIN:DNA replication protein dnad |
| LCRIS_01169 | asnS | 1168843 | 1170141 | 1299 | - | PROTEIN:asparaginyl-tRNA synthetase |
| LCRIS_01170 | LCRIS_01170 | 1170195 | 1170692 | 498 | - | PROTEIN:hypothetical protein |
| LCRIS_01171 | dinG | 1170695 | 1173475 | 2781 | - | PROTEIN:ATP-dependent DNA helicase ding |
| LCRIS_01172 | LCRIS_01172 | 1173491 | 1177099 | 3609 | - | PROTEIN:ATP-dependent helicase/nuclease subunit a |
| LCRIS_01173 | LCRIS_01173 | 1177102 | 1180581 | 3480 | - | PROTEIN:ATP-dependent helicase/nuclease subunit b |
| LCRIS_01174 | mvaK1 | 1180660 | 1181226 | 567 | + | PROTEIN:mevalonate kinase |
| LCRIS_01175 | LCRIS_01175 | 1181235 | 1181567 | 333 | + | PROTEIN:hypothetical protein |
| LCRIS_01176 | mvaD | 1181567 | 1182529 | 963 | + | PROTEIN:diphosphomevalonate decarboxylase |
| LCRIS_01177 | mvaK2 | 1182572 | 1183654 | 1083 | + | PROTEIN:phosphomevalonate kinase |
| LCRIS_01178 | idi | 1183666 | 1184682 | 1017 | + | PROTEIN:isopentenyl-diphosphate delta-isomerase |
| LCRIS_01179 | LCRIS_01179 | 1184735 | 1185145 | 411 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_01180 | LCRIS_01180 | 1185154 | 1185525 | 372 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_01181 | LCRIS_01181 | 1185522 | 1186895 | 1374 | - | PROTEIN:tRNA and rRNA cytosine-c5-methylase |
| LCRIS_01182 | LCRIS_01182 | 1186876 | 1187790 | 915 | - | PROTEIN:hypothetical protein |
| LCRIS_01183 | LCRIS_01183 | 1187843 | 1188595 | 753 | - | PROTEIN:sam-dependent methyltransferase |
| LCRIS_01184 | lepB1 | 1188678 | 1189241 | 564 | + | PROTEIN:signal peptidase i |
| LCRIS_01185 | LCRIS_01185 | 1189312 | 1189500 | 189 | + | PROTEIN:hypothetical protein |
| LCRIS_01186 | LCRIS_01186 | 1189560 | 1190378 | 819 | - | PROTEIN:hypothetical protein |
| LCRIS_01187 | LCRIS_01187 | 1190399 | 1191208 | 810 | - | PROTEIN:hypothetical protein |
| LCRIS_01188 | LCRIS_01188 | 1191218 | 1192048 | 831 | - | PROTEIN:hypothetical protein |
| LCRIS_01189 | LCRIS_01189 | 1192048 | 1192755 | 708 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01190 | gntR2 | 1192748 | 1193122 | 375 | - | PROTEIN:transcriptional regulator, gntr family |
| LCRIS_01191 | pepT2 | 1193429 | 1194670 | 1242 | - | PROTEIN:peptidase t |
| LCRIS_01192 | LCRIS_01192 | 1194689 | 1195486 | 798 | - | PROTEIN:nif3-related protein |
| LCRIS_01193 | LCRIS_01193 | 1195483 | 1196166 | 684 | - | PROTEIN:sam-dependent methyltransferase |
| LCRIS_01194 | LCRIS_01194 | 1196267 | 1196707 | 441 | + | PROTEIN:mutt/nudix family protein |
| LCRIS_01195 | LCRIS_01195 | 1196707 | 1197381 | 675 | + | PROTEIN:enterolysin a |
| LCRIS_01196 | rpoD | 1197428 | 1198540 | 1113 | - | PROTEIN:RNA polymerase sigma factor |
| LCRIS_01197 | dnaG | 1198556 | 1200376 | 1821 | - | PROTEIN:DNA primase |
| LCRIS_01198 | glyS | 1200406 | 1202469 | 2064 | - | PROTEIN:glycyl-tRNA synthetase, beta subunit |
| LCRIS_01199 | glyQ | 1202462 | 1203379 | 918 | - | PROTEIN:glycyl-tRNA synthetase, alpha subunit |
| LCRIS_01200 | recO | 1203639 | 1204394 | 756 | - | PROTEIN:DNA repair protein reco |
| LCRIS_01201 | era | 1205679 | 1206581 | 903 | - | PROTEIN:GTP-binding protein era homolog |
| LCRIS_01202 | LCRIS_01202 | 1206582 | 1207106 | 525 | - | PROTEIN:metalloprotease |
| LCRIS_01203 | phoH | 1207109 | 1208068 | 960 | - | PROTEIN:phoh family protein |
| LCRIS_01204 | LCRIS_01204 | 1208094 | 1208537 | 444 | - | PROTEIN:gatb/yqey domain protein |
| LCRIS_01205 | rpsU | 1208698 | 1208874 | 177 | - | PROTEIN:30S ribosomal protein s21 |
| LCRIS_01206 | LCRIS_01206 | 1209102 | 1209938 | 837 | + | PROTEIN:phosphotransferase |
| LCRIS_01207 | cas1 | 1210016 | 1210912 | 897 | - | PROTEIN:CRISPR-associated protein, cas2 |
| LCRIS_01208 | cas2 | 1210909 | 1211856 | 948 | - | PROTEIN:CRISPR-associated protein, cas1 |
| LCRIS_01209 | LCRIS_01209 | 1211861 | 1212517 | 657 | - | PROTEIN:CRISPR-associated protein, ct1974 |
| LCRIS_01210 | LCRIS_01210 | 1212527 | 1213252 | 726 | - | PROTEIN:CRISPR-associated protein, cas5e-type |
| LCRIS_01211 | casE | 1213233 | 1214342 | 1110 | - | PROTEIN:CRISPR-associated protein, ct1975 |
| LCRIS_01212 | LCRIS_01212 | 1214370 | 1214975 | 606 | - | PROTEIN:CRISPR-associated protein, cse2 |
| LCRIS_01213 | LCRIS_01213 | 1214986 | 1216695 | 1710 | - | PROTEIN:CRISPR-associated protein, cse1 |
| LCRIS_01214 | cas3 | 1216891 | 1219641 | 2751 | - | PROTEIN:CRISPR-associated helicase, cas3 |
| LCRIS_01215 | LCRIS_01215 | 1219697 | 1219969 | 273 | - | PROTEIN:hypothetical protein |
| LCRIS_01216 | LCRIS_01216 | 1220413 | 1220574 | 162 | + | PROTEIN:hypothetical protein |
| LCRIS_01217 | LCRIS_01217 | 1220876 | 1221475 | 600 | + | PROTEIN:transposase (issmu2) |
| LCRIS_01218 | fnr | 1221875 | 1222510 | 636 | - | PROTEIN:transcriptional regulator |
| LCRIS_01219 | LCRIS_01219 | 1222530 | 1223075 | 546 | - | PROTEIN:DNA-binding ferritin-like protein (oxidati |
| LCRIS_01220 | LCRIS_01220 | 1223075 | 1223305 | 231 | - | PROTEIN:copper chaperone |
| LCRIS_01221 | cadA | 1223388 | 1225232 | 1845 | - | PROTEIN:cadmium-transporting atpase |
| LCRIS_01222 | LCRIS_01222 | 1225879 | 1226436 | 558 | - | PROTEIN:hypothetical protein |
| LCRIS_01223 | LCRIS_01223 | 1227232 | 1227441 | 210 | + | PROTEIN:hypothetical protein |
| LCRIS_01224 | LCRIS_01224 | 1228163 | 1228786 | 624 | + | PROTEIN:enterolysin a |
| LCRIS_01225 | msrA | 1228857 | 1229423 | 567 | - | PROTEIN:peptide methionine sulfoxide reductase |
| LCRIS_01226 | LCRIS_01226 | 1229540 | 1229707 | 168 | - | PROTEIN:hypothetical protein |
| LCRIS_01227 | LCRIS_01227 | 1229700 | 1229852 | 153 | - | PROTEIN:hypothetical protein |
| LCRIS_01228 | LCRIS_01228 | 1230532 | 1230954 | 423 | + | PROTEIN:hypothetical protein |
| LCRIS_01229 | LCRIS_01229 | 1231130 | 1232008 | 879 | - | PROTEIN:hypothetical protein |
| LCRIS_01230 | LCRIS_01230 | 1232182 | 1232829 | 648 | + | PROTEIN:temperature-sensitive replication protein |
| LCRIS_01231 | thrB | 1232857 | 1233720 | 864 | - | PROTEIN:homoserine kinase |
| LCRIS_01232 | hom | 1233737 | 1234972 | 1236 | - | PROTEIN:homoserine dehydrogenase |
| LCRIS_01233 | thrC | 1235084 | 1236574 | 1491 | + | PROTEIN:threonine synthase |
| LCRIS_01234 | lysC2 | 1236594 | 1237961 | 1368 | + | PROTEIN:aspartokinase |
| LCRIS_01235 | LCRIS_01235 | 1238052 | 1238612 | 561 | - | PROTEIN:phosphoglycerate mutase family protein |
| LCRIS_01236 | LCRIS_01236 | 1238785 | 1241100 | 2316 | + | PROTEIN:phospholipase d/transphosphatidylase |
| LCRIS_01237 | LCRIS_01237 | 1241204 | 1241671 | 468 | - | PROTEIN:hypothetical protein |
| LCRIS_01238 | LCRIS_01238 | 1241825 | 1243408 | 1584 | - | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_01239 | LCRIS_01239 | 1243405 | 1245003 | 1599 | - | PROTEIN:lactococcin a ABC transporter, ATP binding |
| LCRIS_01240 | LCRIS_01240 | 1245177 | 1246214 | 1038 | - | PROTEIN:hypothetical protein |
| LCRIS_01241 | LCRIS_01241 | 1246785 | 1247447 | 663 | - | PROTEIN:hypothetical protein |
| LCRIS_01242 | LCRIS_01242 | 1247543 | 1248091 | 549 | - | PROTEIN:hypothetical protein |
| LCRIS_01243 | LCRIS_01243 | 1248182 | 1248799 | 618 | - | PROTEIN:transcriptional regulator |
| LCRIS_01244 | LCRIS_01244 | 1248789 | 1248950 | 162 | - | PROTEIN:hypothetical protein |
| LCRIS_01245 | LCRIS_01245 | 1248966 | 1250132 | 1167 | - | PROTEIN:acetyltransferase |
| LCRIS_01246 | citX | 1250248 | 1250784 | 537 | - | PROTEIN:apo-citrate lyase |
| LCRIS_01247 | arcC | 1250798 | 1251403 | 606 | - | PROTEIN:ornithine carbamoyltransferase |
| LCRIS_01248 | LCRIS_01248 | 1251412 | 1251591 | 180 | - | PROTEIN:aspartate/ornithine carbamoyltransferase |
| LCRIS_01249 | argF | 1251624 | 1251983 | 360 | - | PROTEIN:ornithine carbamoyltransferase |
| LCRIS_01250 | arcA | 1251994 | 1253223 | 1230 | - | PROTEIN:arginine deiminase |
| LCRIS_01251 | LCRIS_01251 | 1253297 | 1253650 | 354 | - | PROTEIN:transcription regulatory protein |
| LCRIS_01252 | LCRIS_01252 | 1253640 | 1254146 | 507 | - | PROTEIN:transcriptional regulator, lysr family (n- |
| LCRIS_01253 | LCRIS_01253 | 1254275 | 1256548 | 2274 | + | PROTEIN:cation-transporting atpase |
| LCRIS_01254 | LCRIS_01254 | 1256616 | 1256930 | 315 | - | PROTEIN:hypothetical protein |
| LCRIS_01255 | LCRIS_01255 | 1257018 | 1257287 | 270 | - | PROTEIN:hypothetical protein |
| LCRIS_01256 | LCRIS_01256 | 1257311 | 1257475 | 165 | - | PROTEIN:hypothetical protein |
| LCRIS_01257 | LCRIS_01257 | 1257736 | 1258830 | 1095 | + | PROTEIN:surface protein |
| LCRIS_01258 | merA | 1258874 | 1259164 | 291 | - | PROTEIN:mercuric reductase |
| LCRIS_01259 | LCRIS_01259 | 1259161 | 1260183 | 1023 | - | PROTEIN:oxidoreductase protein |
| LCRIS_01260 | apt | 1260636 | 1261163 | 528 | - | PROTEIN:adenine phosphoribosyltransferase |
| LCRIS_01261 | recJ | 1261268 | 1263541 | 2274 | - | PROTEIN:single-stranded-DNA-specific exonuclease r |
| LCRIS_01262 | srtA | 1263662 | 1264300 | 639 | - | PROTEIN:sortase |
| LCRIS_01263 | lepA | 1264348 | 1266186 | 1839 | - | PROTEIN:GTP-binding protein lepa |
| LCRIS_01264 | LCRIS_01264 | 1266294 | 1266749 | 456 | + | PROTEIN:nudix family hydrolase |
| LCRIS_01265 | dnaJ | 1266804 | 1267955 | 1152 | - | PROTEIN:chaperone protein dnaj |
| LCRIS_01266 | dnaK | 1268038 | 1269891 | 1854 | - | PROTEIN:chaperone protein dnak |
| LCRIS_01267 | grpE | 1269908 | 1270492 | 585 | - | PROTEIN:protein grpe |
| LCRIS_01268 | hrcA | 1270505 | 1271554 | 1050 | - | PROTEIN:heat-inducible transcription repressor hrc |
| LCRIS_01269 | ribF | 1271692 | 1272642 | 951 | - | PROTEIN:riboflavin kinase / fmn adenylyltransferas |
| LCRIS_01270 | truB | 1272668 | 1273540 | 873 | - | PROTEIN:tRNA pseudouridine synthase b |
| LCRIS_01271 | rbfA | 1273596 | 1273961 | 366 | - | PROTEIN:ribosome-binding factor a |
| LCRIS_01272 | infB | 1273981 | 1276584 | 2604 | - | PROTEIN:translation initiation factor if-2 |
| LCRIS_01273 | LCRIS_01273 | 1276589 | 1276900 | 312 | - | PROTEIN:ribosomal protein l7a family |
| LCRIS_01274 | LCRIS_01274 | 1276903 | 1277199 | 297 | - | PROTEIN:nucleic-acid-binding protein implicated in |
| LCRIS_01275 | nusA | 1277209 | 1278402 | 1194 | - | PROTEIN:transcription termination-antitermination |
| LCRIS_01276 | LCRIS_01276 | 1278422 | 1278898 | 477 | - | PROTEIN:hypothetical protein |
| LCRIS_01277 | polC | 1279009 | 1283319 | 4311 | - | PROTEIN:DNA polymerase iii, alpha subunit |
| LCRIS_01278 | proS | 1283325 | 1285022 | 1698 | - | PROTEIN:prolyl-tRNA synthetase |
| LCRIS_01279 | LCRIS_01279 | 1285050 | 1286306 | 1257 | - | PROTEIN:membrane-associated zinc metalloprotease |
| LCRIS_01280 | cdsA | 1286319 | 1287059 | 741 | - | PROTEIN:phosphatidate cytidylyltransferase |
| LCRIS_01281 | uppS | 1287137 | 1287871 | 735 | - | PROTEIN:undecaprenyl pyrophosphate synthetase |
| LCRIS_01282 | frr | 1287874 | 1288431 | 558 | - | PROTEIN:ribosome-recycling factor |
| LCRIS_01283 | pyrH | 1288431 | 1289156 | 726 | - | PROTEIN:uridylate kinase |
| LCRIS_01284 | tsf | 1289301 | 1290326 | 1026 | - | PROTEIN:elongation factor ts |
| LCRIS_01285 | rpsB | 1290360 | 1291133 | 774 | - | PROTEIN:30S ribosomal protein s2 |
| LCRIS_01286 | LCRIS_01286 | 1291297 | 1292328 | 1032 | - | PROTEIN:o-methyltransferase |
| LCRIS_01287 | plsC | 1292402 | 1293016 | 615 | + | PROTEIN:1-acyl-sn-glycerol-3-phosphate acyltransfe |
| LCRIS_01288 | cfa | 1293129 | 1294307 | 1179 | + | PROTEIN:cyclopropane-fatty-acyl-phospholipid synth |
| LCRIS_01289 | pepO2 | 1294367 | 1296310 | 1944 | - | PROTEIN:endopeptidase o |
| LCRIS_01290 | mdlB | 1296403 | 1298202 | 1800 | - | PROTEIN:ABC transporter, ATP-binding/permease prot |
| LCRIS_01291 | mdlA | 1298195 | 1299961 | 1767 | - | PROTEIN:ABC transporter, ATP-binding/permease prot |
| LCRIS_01292 | LCRIS_01292 | 1300042 | 1300260 | 219 | - | PROTEIN:hypothetical protein |
| LCRIS_01293 | LCRIS_01293 | 1300330 | 1300596 | 267 | - | PROTEIN:hypothetical protein |
| LCRIS_01294 | lexA | 1300753 | 1301379 | 627 | + | PROTEIN:lexa repressor |
| LCRIS_01295 | LCRIS_01295 | 1301418 | 1302197 | 780 | - | PROTEIN:acyl-CoA thioesterase |
| LCRIS_01296 | dpo | 1302190 | 1302849 | 660 | - | PROTEIN:uracil-DNA glycosylase family protein |
| LCRIS_01297 | LCRIS_01297 | 1303199 | 1304323 | 1125 | - | PROTEIN:hypothetical protein |
| LCRIS_01298 | rplS | 1304435 | 1304782 | 348 | - | PROTEIN:50S ribosomal protein l19 |
| LCRIS_01299 | trmD | 1304896 | 1305615 | 720 | - | PROTEIN:tRNA (guanine-n(1)-)-methyltransferase |
| LCRIS_01300 | rimM | 1305605 | 1306120 | 516 | - | PROTEIN:ribosome maturation factor rimm |
| LCRIS_01301 | rpsP | 1306187 | 1306459 | 273 | - | PROTEIN:30S ribosomal protein s16 |
| LCRIS_01302 | ffh | 1306547 | 1307977 | 1431 | - | PROTEIN:signal recognition particle protein ffh |
| LCRIS_01303 | LCRIS_01303 | 1307981 | 1308322 | 342 | - | PROTEIN:hypothetical protein |
| LCRIS_01304 | LCRIS_01304 | 1308495 | 1309925 | 1431 | + | PROTEIN:amino acid transporter |
| LCRIS_01305 | LCRIS_01305 | 1309982 | 1311808 | 1827 | - | PROTEIN:ABC-type antimicrobial peptide transport s |
| LCRIS_01306 | LCRIS_01306 | 1311823 | 1312569 | 747 | - | PROTEIN:ABC-type antimicrobial peptide transport s |
| LCRIS_01307 | pepD1 | 1313116 | 1314537 | 1422 | - | PROTEIN:dipeptidase |
| LCRIS_01308 | ftsY | 1314582 | 1315823 | 1242 | - | PROTEIN:cell division protein, signal recognition |
| LCRIS_01309 | smc | 1315824 | 1319393 | 3570 | - | PROTEIN:chromosome segregation protein smc |
| LCRIS_01310 | rnc | 1319406 | 1320095 | 690 | - | PROTEIN:ribonuclease iii |
| LCRIS_01311 | oppA2 | 1320201 | 1321952 | 1752 | - | PROTEIN:oligopeptide ABC transporter, oligopeptide |
| LCRIS_01312 | oppC2 | 1322266 | 1323045 | 780 | - | PROTEIN:oligopeptide ABC transporter, permease pro |
| LCRIS_01313 | oppB2 | 1323059 | 1324018 | 960 | - | PROTEIN:oligopeptide ABC transporter, permease pro |
| LCRIS_01314 | oppF2 | 1324018 | 1324986 | 969 | - | PROTEIN:oligopeptide ABC transporter, ATP-binding |
| LCRIS_01315 | oppD2 | 1324990 | 1326006 | 1017 | - | PROTEIN:oligopeptide ABC transporter, ATP-binding |
| LCRIS_01316 | acpP | 1326125 | 1326367 | 243 | - | PROTEIN:acyl carrier protein |
| LCRIS_01317 | plsX | 1326398 | 1327399 | 1002 | - | PROTEIN:fatty acid/phospholipid synthesis protein |
| LCRIS_01318 | recG | 1327419 | 1329455 | 2037 | - | PROTEIN:ATP-dependent DNA helicase recg |
| LCRIS_01319 | LCRIS_01319 | 1329458 | 1331119 | 1662 | - | PROTEIN:dihydroacetone kinase |
| LCRIS_01320 | LCRIS_01320 | 1331141 | 1331503 | 363 | - | PROTEIN:alkaline shock protein |
| LCRIS_01321 | rpmB | 1331671 | 1331841 | 171 | + | PROTEIN:50S ribosomal protein l28 |
| LCRIS_01322 | LCRIS_01322 | 1332089 | 1332703 | 615 | + | PROTEIN:transposase |
| LCRIS_01323 | LCRIS_01323 | 1332733 | 1333368 | 636 | + | PROTEIN:transposase |
| LCRIS_01324 | LCRIS_01324 | 1333456 | 1333572 | 117 | - | PROTEIN:hypothetical protein |
| LCRIS_01325 | thiN | 1333799 | 1334473 | 675 | - | PROTEIN:thiamine pyrophosphokinase |
| LCRIS_01326 | rpe1 | 1334473 | 1335123 | 651 | - | PROTEIN:ribulose-phosphate 3-epimerase |
| LCRIS_01327 | rsgA | 1335136 | 1336026 | 891 | - | PROTEIN:ribosome biogenesis GTPase rsga |
| LCRIS_01328 | prkC | 1336027 | 1338057 | 2031 | - | PROTEIN:serine/threonine protein kinase |
| LCRIS_01329 | LCRIS_01329 | 1338047 | 1338802 | 756 | - | PROTEIN:serine/threonine protein phosphatase |
| LCRIS_01330 | sun | 1338807 | 1340132 | 1326 | - | PROTEIN:sun protein |
| LCRIS_01331 | fmt | 1340119 | 1341063 | 945 | - | PROTEIN:methionyl-tRNA formyltransferase |
| LCRIS_01332 | priA | 1341076 | 1343475 | 2400 | - | PROTEIN:primosomal protein n' |
| LCRIS_01333 | rpoZ | 1343526 | 1343750 | 225 | - | PROTEIN:DNA-directed RNA polymerase, omega subunit |
| LCRIS_01334 | gmk1 | 1343756 | 1344370 | 615 | - | PROTEIN:guanylate kinase |
| LCRIS_01335 | LCRIS_01335 | 1344478 | 1344915 | 438 | - | PROTEIN:cytoplasmic protein |
| LCRIS_01336 | recN | 1344908 | 1346590 | 1683 | - | PROTEIN:DNA repair protein recn |
| LCRIS_01337 | tlyA | 1346601 | 1347413 | 813 | - | PROTEIN:hemolysin a |
| LCRIS_01338 | ispA | 1347414 | 1348283 | 870 | - | PROTEIN:geranyltranstransferase |
| LCRIS_01339 | xseB | 1348286 | 1348528 | 243 | - | PROTEIN:exodeoxyribonuclease vii, small subunit |
| LCRIS_01340 | xseA | 1348509 | 1349876 | 1368 | - | PROTEIN:exodeoxyribonuclease vii, large subunit |
| LCRIS_01341 | folD | 1349863 | 1350714 | 852 | - | PROTEIN:bifunctional protein fold |
| LCRIS_01342 | nusB | 1350771 | 1351166 | 396 | - | PROTEIN:transcription termination factor |
| LCRIS_01343 | LCRIS_01343 | 1351166 | 1351597 | 432 | - | PROTEIN:alkaline shock protein |
| LCRIS_01344 | efp1 | 1351616 | 1352185 | 570 | - | PROTEIN:elongation factor p |
| LCRIS_01345 | pepP1 | 1352266 | 1353375 | 1110 | - | PROTEIN:xaa-pro dipeptidase |
| LCRIS_01346 | rpmA | 1353492 | 1353782 | 291 | - | PROTEIN:50S ribosomal protein l27 |
| LCRIS_01347 | rplU | 1353801 | 1354112 | 312 | - | PROTEIN:50S ribosomal protein l21 |
| LCRIS_01348 | LCRIS_01348 | 1354271 | 1355107 | 837 | - | PROTEIN:hypothetical protein |
| LCRIS_01349 | LCRIS_01349 | 1355191 | 1356567 | 1377 | - | PROTEIN:v-type sodium ATP synthase subunit |
| LCRIS_01350 | LCRIS_01350 | 1356725 | 1357123 | 399 | - | PROTEIN:hypothetical protein |
| LCRIS_01351 | LCRIS_01351 | 1357261 | 1357440 | 180 | - | PROTEIN:4-oxalocrotonate tautomerase |
| LCRIS_01352 | LCRIS_01352 | 1357489 | 1359282 | 1794 | - | PROTEIN:hypothetical protein |
| LCRIS_01353 | LCRIS_01353 | 1359286 | 1360191 | 906 | - | PROTEIN:alpha-beta superfamily hydrolase |
| LCRIS_01354 | LCRIS_01354 | 1360321 | 1361559 | 1239 | - | PROTEIN:lysin |
| LCRIS_02095 | tRNA-Arg | 1361822 | 1361895 | 74 | - | RNA:Arg tRNA |
| LCRIS_01355 | LCRIS_01355 | 1362052 | 1362948 | 897 | + | PROTEIN:lipase |
| LCRIS_01356 | LCRIS_01356 | 1362961 | 1363317 | 357 | + | PROTEIN:arsenate reductase |
| LCRIS_01357 | LCRIS_01357 | 1363363 | 1364943 | 1581 | - | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_01358 | LCRIS_01358 | 1364943 | 1366523 | 1581 | - | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_01359 | LCRIS_01359 | 1366857 | 1367672 | 816 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_01360 | LCRIS_01360 | 1367732 | 1368058 | 327 | - | PROTEIN:hypothetical protein |
| LCRIS_01361 | LCRIS_01361 | 1368180 | 1368440 | 261 | - | PROTEIN:hypothetical protein |
| LCRIS_01362 | LCRIS_01362 | 1368563 | 1368829 | 267 | - | PROTEIN:alpha/beta hydrolase superfamily protein |
| LCRIS_01363 | LCRIS_01363 | 1368956 | 1369195 | 240 | + | PROTEIN:aminopeptidase |
| LCRIS_01364 | pepXP | 1369290 | 1371338 | 2049 | + | PROTEIN:xaa-pro dipeptidyl-peptidase |
| LCRIS_01365 | msrB | 1371395 | 1371832 | 438 | - | PROTEIN:peptide methionine sulfoxide reductase |
| LCRIS_01366 | LCRIS_01366 | 1371848 | 1372129 | 282 | - | PROTEIN:hypothetical protein |
| LCRIS_01367 | LCRIS_01367 | 1372255 | 1373118 | 864 | - | PROTEIN:glucose uptake permease |
| LCRIS_01368 | lctP1 | 1373263 | 1374789 | 1527 | - | PROTEIN:L-lactate permease |
| LCRIS_01369 | carB2 | 1375053 | 1378241 | 3189 | - | PROTEIN:carbamoyl-phosphate synthase, large subuni |
| LCRIS_01370 | carA2 | 1378234 | 1379319 | 1086 | - | PROTEIN:carbamoyl-phosphate synthase, small subuni |
| LCRIS_01371 | pyrC | 1379319 | 1380596 | 1278 | - | PROTEIN:dihydroorotase |
| LCRIS_01372 | pyrB | 1380596 | 1381552 | 957 | - | PROTEIN:aspartate carbamoyltransferase |
| LCRIS_01373 | pyrR2 | 1381693 | 1382235 | 543 | - | PROTEIN:uracil phosphoribosyltransferase / pyrimid |
| LCRIS_01374 | pyrD | 1382404 | 1383327 | 924 | - | PROTEIN:dihydroorotate dehydrogenase |
| LCRIS_01375 | pyrF | 1383578 | 1384282 | 705 | + | PROTEIN:orotidine-5'-phosphate decarboxylase |
| LCRIS_01376 | pyrE | 1384284 | 1384922 | 639 | + | PROTEIN:orotate phosphoribosyltransferase |
| LCRIS_01377 | LCRIS_01377 | 1385062 | 1385520 | 459 | + | PROTEIN:chloride channel protein |
| LCRIS_01378 | LCRIS_01378 | 1385791 | 1387293 | 1503 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01379 | LCRIS_01379 | 1387346 | 1387639 | 294 | - | PROTEIN:hypothetical protein |
| LCRIS_01380 | sdaB | 1387768 | 1388430 | 663 | + | PROTEIN:L-serine dehydratase, beta subunit |
| LCRIS_01381 | sdaA | 1388445 | 1389326 | 882 | + | PROTEIN:L-serine dehydratase, alpha subunit |
| LCRIS_01382 | LCRIS_01382 | 1389335 | 1389649 | 315 | + | PROTEIN:metal-sulfur cluster biosynthetic enzyme |
| LCRIS_01383 | dapE1 | 1389697 | 1390362 | 666 | - | PROTEIN:succinyl-diaminopimelate desuccinylase |
| LCRIS_01384 | dapE2 | 1390352 | 1390636 | 285 | - | PROTEIN:succinyl-diaminopimelate desuccinylase |
| LCRIS_01385 | LCRIS_01385 | 1390844 | 1392127 | 1284 | - | PROTEIN:transposase |
| LCRIS_01386 | LCRIS_01386 | 1392124 | 1392231 | 108 | - | PROTEIN:hypothetical protein |
| LCRIS_01387 | LCRIS_01387 | 1392270 | 1392746 | 477 | - | PROTEIN:first orf in partial transposon isc1913 |
| LCRIS_01388 | lctO | 1393098 | 1394378 | 1281 | - | PROTEIN:L-lactate oxidase |
| LCRIS_01389 | ackA2 | 1394663 | 1395862 | 1200 | + | PROTEIN:acetate kinase |
| LCRIS_01390 | LCRIS_01390 | 1395840 | 1397573 | 1734 | - | PROTEIN:transposase |
| LCRIS_01391 | LCRIS_01391 | 1397728 | 1399224 | 1497 | - | PROTEIN:amino acid permease |
| LCRIS_01392 | LCRIS_01392 | 1399503 | 1401209 | 1707 | - | PROTEIN:hypothetical protein |
| LCRIS_01393 | LCRIS_01393 | 1401378 | 1402100 | 723 | - | PROTEIN:orfb |
| LCRIS_01394 | LCRIS_01394 | 1402195 | 1403694 | 1500 | - | PROTEIN:hypothetical protein |
| LCRIS_01395 | LCRIS_01395 | 1403840 | 1404589 | 750 | - | PROTEIN:hypothetical protein |
| LCRIS_01396 | LCRIS_01396 | 1404641 | 1405153 | 513 | - | PROTEIN:hypothetical protein |
| LCRIS_01397 | LCRIS_01397 | 1405157 | 1405561 | 405 | - | PROTEIN:redox protein, regulator of disulfide bond |
| LCRIS_01398 | LCRIS_01398 | 1405571 | 1406743 | 1173 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01399 | LCRIS_01399 | 1406791 | 1408068 | 1278 | - | PROTEIN:sensor protein |
| LCRIS_01400 | LCRIS_01400 | 1408201 | 1408743 | 543 | - | PROTEIN:response regulator |
| LCRIS_01401 | LCRIS_01401 | 1408750 | 1409364 | 615 | - | PROTEIN:GTP pyrophosphokinase |
| LCRIS_01402 | gtfA2 | 1409536 | 1410978 | 1443 | - | PROTEIN:sucrose phosphorylase |
| LCRIS_01403 | galA1 | 1410990 | 1413191 | 2202 | - | PROTEIN:alpha-galactosidase |
| LCRIS_01404 | msmK2 | 1413201 | 1414313 | 1113 | - | PROTEIN:sugar ABC transporter, ATP-binding protein |
| LCRIS_01405 | msmG | 1414340 | 1415173 | 834 | - | PROTEIN:sugar ABC transporter, permease protein |
| LCRIS_01406 | msmF | 1415188 | 1416063 | 876 | - | PROTEIN:sugar ABC transporter, permease protein |
| LCRIS_01407 | msmE | 1416076 | 1417332 | 1257 | - | PROTEIN:sugar ABC transporter, sugar-binding prote |
| LCRIS_01408 | LCRIS_01408 | 1417519 | 1418475 | 957 | + | PROTEIN:sucrose operon repressor scrr |
| LCRIS_01409 | LCRIS_01409 | 1418532 | 1418765 | 234 | - | PROTEIN:hypothetical protein |
| LCRIS_01410 | galM | 1418898 | 1419893 | 996 | - | PROTEIN:aldose 1-epimerase |
| LCRIS_01411 | galT | 1420016 | 1421485 | 1470 | - | PROTEIN:galactose-1-phosphate uridylyltransferase |
| LCRIS_01412 | galK | 1421504 | 1422667 | 1164 | - | PROTEIN:galactokinase |
| LCRIS_01413 | lacZ | 1423035 | 1425041 | 2007 | - | PROTEIN:beta-galactosidase |
| LCRIS_01414 | LCRIS_01414 | 1425052 | 1426971 | 1920 | - | PROTEIN:lactose permease |
| LCRIS_01415 | lacR | 1427164 | 1428171 | 1008 | - | PROTEIN:transcription repressor of beta-galactosid |
| LCRIS_01416 | lacL | 1428396 | 1430276 | 1881 | + | PROTEIN:beta-galactosidase large subunit |
| LCRIS_01417 | lacM | 1430260 | 1431210 | 951 | + | PROTEIN:beta-galactosidase small subunit |
| LCRIS_01418 | galE | 1431313 | 1432305 | 993 | + | PROTEIN:udp-glucose 4-epimerase |
| LCRIS_01419 | LCRIS_01419 | 1432415 | 1433290 | 876 | - | PROTEIN:abortive infection protein |
| LCRIS_01420 | LCRIS_01420 | 1433287 | 1433877 | 591 | - | PROTEIN:abortive infection protein abigi |
| LCRIS_01421 | galR | 1434232 | 1435263 | 1032 | + | PROTEIN:galr |
| LCRIS_01422 | LCRIS_01422 | 1435901 | 1438279 | 2379 | - | PROTEIN:hypothetical protein |
| LCRIS_01423 | LCRIS_01423 | 1438545 | 1439771 | 1227 | + | PROTEIN:transposase |
| LCRIS_01424 | malY | 1440041 | 1441222 | 1182 | - | PROTEIN:maltose regulon modulator maly |
| LCRIS_01425 | pepP2 | 1441230 | 1442306 | 1077 | - | PROTEIN:xaa-pro dipeptidase |
| LCRIS_01426 | LCRIS_01426 | 1442299 | 1442913 | 615 | - | PROTEIN:hypothetical protein |
| LCRIS_01427 | lacE | 1442936 | 1444252 | 1317 | - | PROTEIN:PTS system lactose/cellobiose-specific iic |
| LCRIS_01428 | celA2 | 1444255 | 1444599 | 345 | - | PROTEIN:PTS system lactose/cellobiose-specific iib |
| LCRIS_01429 | celC2 | 1444618 | 1444941 | 324 | - | PROTEIN:PTS system lactose/cellobiose-specific iia |
| LCRIS_01430 | ypdE | 1444956 | 1445999 | 1044 | - | PROTEIN:aminopeptidase |
| LCRIS_01431 | licR | 1446002 | 1447897 | 1896 | - | PROTEIN:PTS system iia component |
| LCRIS_01432 | LCRIS_01432 | 1447984 | 1448775 | 792 | - | PROTEIN:short-chain dehydrogenase/reductase sdr |
| LCRIS_01433 | LCRIS_01433 | 1448812 | 1449687 | 876 | - | PROTEIN:short-chain dehydrogenase/reductase sdr |
| LCRIS_01434 | LCRIS_01434 | 1449706 | 1450350 | 645 | - | PROTEIN:nitroreductase |
| LCRIS_01435 | LCRIS_01435 | 1450726 | 1451118 | 393 | + | PROTEIN:truncated transposase |
| LCRIS_01436 | manX1 | 1451345 | 1451725 | 381 | - | PROTEIN:PTS system mannose/fructose-specific iia c |
| LCRIS_01437 | manZ2 | 1451740 | 1452567 | 828 | - | PROTEIN:PTS system mannose/fructose-specific iid c |
| LCRIS_01438 | manY2 | 1452545 | 1453333 | 789 | - | PROTEIN:PTS system iic component |
| LCRIS_01439 | manX2 | 1453350 | 1453814 | 465 | - | PROTEIN:PTS system mannose/fructose-specific iib c |
| LCRIS_01440 | LCRIS_01440 | 1453825 | 1454532 | 708 | - | PROTEIN:transcriptional regulator, gntr family |
| LCRIS_01441 | LCRIS_01441 | 1454552 | 1455523 | 972 | - | PROTEIN:sugar isomerase |
| LCRIS_01442 | LCRIS_01442 | 1455712 | 1456761 | 1050 | - | PROTEIN:transposase |
| LCRIS_01443 | malE1 | 1456874 | 1458091 | 1218 | - | PROTEIN:maltose/maltodextrin ABC transporter, malt |
| LCRIS_01444 | LCRIS_01444 | 1458095 | 1459075 | 981 | - | PROTEIN:transcriptional regulator, repressor of su |
| LCRIS_01445 | LCRIS_01445 | 1459086 | 1461293 | 2208 | - | PROTEIN:glycosyl hydrolase, family 31 |
| LCRIS_01446 | umuC | 1461525 | 1462844 | 1320 | + | PROTEIN:nucleotidyltransferase/DNA polymerase for |
| LCRIS_01447 | LCRIS_01447 | 1462837 | 1463205 | 369 | + | PROTEIN:hypothetical protein |
| LCRIS_01448 | LCRIS_01448 | 1463447 | 1464727 | 1281 | + | PROTEIN:transposase |
| LCRIS_01449 | LCRIS_01449 | 1464802 | 1465791 | 990 | - | PROTEIN:hypothetical protein |
| LCRIS_01450 | LCRIS_01450 | 1465936 | 1466850 | 915 | - | PROTEIN:ABC transporter |
| LCRIS_01451 | glnA | 1467140 | 1468477 | 1338 | - | PROTEIN:glutamine synthetase |
| LCRIS_01452 | LCRIS_01452 | 1468665 | 1469864 | 1200 | - | PROTEIN:aluminum resistance protein |
| LCRIS_01453 | miaA | 1469857 | 1470777 | 921 | - | PROTEIN:tRNA delta(2)-isopentenylpyrophosphate tra |
| LCRIS_01454 | LCRIS_01454 | 1470855 | 1471256 | 402 | - | PROTEIN:rhodanese-related sulfurtransferase |
| LCRIS_01455 | LCRIS_01455 | 1471315 | 1471554 | 240 | - | PROTEIN:cytosolic protein |
| LCRIS_01456 | LCRIS_01456 | 1471554 | 1472240 | 687 | - | PROTEIN:hypothetical protein |
| LCRIS_01457 | LCRIS_01457 | 1472274 | 1472870 | 597 | - | PROTEIN:5-formyltetrahydrofolate cyclo-ligase |
| LCRIS_01458 | rpmG2 | 1472892 | 1473050 | 159 | - | PROTEIN:50S ribosomal protein l33 |
| LCRIS_01459 | pbp2 | 1473117 | 1475225 | 2109 | - | PROTEIN:penicillin-binding protein |
| LCRIS_01460 | LCRIS_01460 | 1475316 | 1477952 | 2637 | + | PROTEIN:ABC transporter, permease protein |
| LCRIS_01461 | LCRIS_01461 | 1478015 | 1478938 | 924 | + | PROTEIN:N-acetylglucosamine kinase |
| LCRIS_01462 | LCRIS_01462 | 1479206 | 1479454 | 249 | + | PROTEIN:addiction module antitoxin, relb/dinj fami |
| LCRIS_01463 | LCRIS_01463 | 1479457 | 1479750 | 294 | + | PROTEIN:hypothetical protein |
| LCRIS_01464 | hupB2 | 1480509 | 1480793 | 285 | + | PROTEIN:DNA-binding protein hu |
| LCRIS_01465 | fepC | 1480838 | 1481611 | 774 | - | PROTEIN:iron compound ABC transporter, ATP-binding |
| LCRIS_01466 | LCRIS_01466 | 1481604 | 1482530 | 927 | - | PROTEIN:iron compound ABC transporter, permease co |
| LCRIS_01467 | LCRIS_01467 | 1482536 | 1483417 | 882 | - | PROTEIN:iron compound ABC transporter, iron compou |
| LCRIS_01468 | LCRIS_01468 | 1483414 | 1484133 | 720 | - | PROTEIN:hypothetical protein |
| LCRIS_01469 | LCRIS_01469 | 1484133 | 1484732 | 600 | - | PROTEIN:hypothetical protein |
| LCRIS_01470 | LCRIS_01470 | 1484848 | 1485441 | 594 | - | PROTEIN:hypothetical protein |
| LCRIS_01471 | LCRIS_01471 | 1485646 | 1486425 | 780 | + | PROTEIN:s-layer protein |
| LCRIS_01472 | LCRIS_01472 | 1486510 | 1486755 | 246 | - | PROTEIN:hypothetical protein |
| LCRIS_01473 | LCRIS_01473 | 1487207 | 1488613 | 1407 | + | PROTEIN:transposase |
| LCRIS_01474 | LCRIS_01474 | 1488711 | 1489202 | 492 | - | PROTEIN:hypothetical protein |
| LCRIS_01475 | LCRIS_01475 | 1489195 | 1490013 | 819 | - | PROTEIN:hypothetical protein |
| LCRIS_01476 | LCRIS_01476 | 1490010 | 1491281 | 1272 | - | PROTEIN:metal-dependent amidase/aminoacylase/carbo |
| LCRIS_01477 | LCRIS_01477 | 1491500 | 1492705 | 1206 | - | PROTEIN:amidohydrolase |
| LCRIS_01478 | oppA3 | 1492716 | 1494347 | 1632 | - | PROTEIN:oligopeptide ABC transporter, substrate bi |
| LCRIS_01479 | LCRIS_01479 | 1494744 | 1495559 | 816 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_01480 | sufI | 1495788 | 1497341 | 1554 | + | PROTEIN:multicopper oxidase |
| LCRIS_01481 | LCRIS_01481 | 1497354 | 1497530 | 177 | + | PROTEIN:hypothetical protein |
| LCRIS_01482 | metQ2 | 1497902 | 1498753 | 852 | + | PROTEIN:ABC-type metal ion transport system, subst |
| LCRIS_01483 | ldh3 | 1498820 | 1499731 | 912 | - | PROTEIN:L-2-hydroxyisocaproate dehydrogenase |
| LCRIS_01484 | greA2 | 1499793 | 1500269 | 477 | - | PROTEIN:transcription elongation factor |
| LCRIS_01485 | pheT | 1500348 | 1502768 | 2421 | - | PROTEIN:phenylalanyl-tRNA synthetase, beta subunit |
| LCRIS_01486 | pheS | 1502772 | 1503821 | 1050 | - | PROTEIN:phenylalanyl-tRNA synthetase, alpha subuni |
| LCRIS_01487 | LCRIS_01487 | 1504105 | 1504974 | 870 | - | PROTEIN:amino acid ABC transporter, amino acid-bin |
| LCRIS_01488 | gltL | 1504981 | 1505721 | 741 | - | PROTEIN:amino acid ABC transporter, ATP-binding pr |
| LCRIS_01489 | glnP3 | 1505731 | 1506402 | 672 | - | PROTEIN:amino acid ABC transporter, permease prote |
| LCRIS_01490 | glnP4 | 1506383 | 1507042 | 660 | - | PROTEIN:amino acid ABC transporter, permease prote |
| LCRIS_01491 | LCRIS_01491 | 1507343 | 1507693 | 351 | - | PROTEIN:transcriptional regulator |
| LCRIS_01492 | spoU | 1507801 | 1508568 | 768 | - | PROTEIN:rRNA methylase |
| LCRIS_01493 | acyP | 1508668 | 1508940 | 273 | + | PROTEIN:acylphosphatase |
| LCRIS_01494 | yidC | 1508984 | 1509949 | 966 | + | PROTEIN:preprotein translocase subunit yidc |
| LCRIS_01495 | LCRIS_01495 | 1509996 | 1510484 | 489 | + | PROTEIN:ybak/prolyl-tRNA synthetase associated reg |
| LCRIS_01496 | LCRIS_01496 | 1510676 | 1511857 | 1182 | - | PROTEIN:s-layer protein |
| LCRIS_01497 | LCRIS_01497 | 1511926 | 1512225 | 300 | - | PROTEIN:abortive infection protein |
| LCRIS_01498 | LCRIS_01498 | 1512332 | 1512526 | 195 | - | PROTEIN:hypothetical protein |
| LCRIS_01499 | LCRIS_01499 | 1512828 | 1514405 | 1578 | - | PROTEIN:sensor protein |
| LCRIS_01500 | LCRIS_01500 | 1514383 | 1515099 | 717 | - | PROTEIN:response regulator |
| LCRIS_01501 | LCRIS_01501 | 1515268 | 1515831 | 564 | - | PROTEIN:metal-binding, possibly nucleic acid-bindi |
| LCRIS_01502 | LCRIS_01502 | 1515834 | 1516985 | 1152 | - | PROTEIN:hypothetical protein |
| LCRIS_01503 | LCRIS_01503 | 1516989 | 1517336 | 348 | - | PROTEIN:iojap family protein |
| LCRIS_01504 | LCRIS_01504 | 1517355 | 1517948 | 594 | - | PROTEIN:nad metabolism hydrolase of hd superfamily |
| LCRIS_01505 | nadD | 1517929 | 1518582 | 654 | - | PROTEIN:nicotinate-nucleotide adenylyltransferase |
| LCRIS_01506 | LCRIS_01506 | 1518593 | 1519702 | 1110 | - | PROTEIN:GTP-binding protein |
| LCRIS_01507 | LCRIS_01507 | 1519695 | 1520219 | 525 | - | PROTEIN:hydrolase, had superfamily |
| LCRIS_01508 | add | 1520357 | 1521361 | 1005 | + | PROTEIN:adenosine deaminase |
| LCRIS_01509 | galA2 | 1521395 | 1523593 | 2199 | - | PROTEIN:alpha-galactosidase |
| LCRIS_01510 | scrA2 | 1523603 | 1525474 | 1872 | - | PROTEIN:PTS system sucrose-specific iiABC componen |
| LCRIS_01511 | LCRIS_01511 | 1525492 | 1525809 | 318 | - | PROTEIN:concanavalin a-like lectin/glucanase |
| LCRIS_01512 | LCRIS_01512 | 1525811 | 1526818 | 1008 | - | PROTEIN:sucrase |
| LCRIS_01513 | LCRIS_01513 | 1526923 | 1527588 | 666 | + | PROTEIN:arac protein, arabinose-binding/dimerisati |
| LCRIS_01514 | LCRIS_01514 | 1527594 | 1527761 | 168 | + | PROTEIN:arac-type DNA-binding domain-containing pr |
| LCRIS_01515 | rplT | 1527791 | 1528147 | 357 | - | PROTEIN:50S ribosomal protein l20 |
| LCRIS_01516 | rpmI | 1528192 | 1528392 | 201 | - | PROTEIN:50S ribosomal protein l35 |
| LCRIS_01517 | infC | 1528414 | 1528863 | 450 | - | PROTEIN:translation initiation factor if-3 |
| LCRIS_01518 | LCRIS_01518 | 1529124 | 1529684 | 561 | - | PROTEIN:streptothricin acetyltransferase |
| LCRIS_01519 | LCRIS_01519 | 1529716 | 1530480 | 765 | - | PROTEIN:ABC-type polysaccharide/polyol phosphate e |
| LCRIS_01520 | LCRIS_01520 | 1530480 | 1531406 | 927 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01521 | LCRIS_01521 | 1531409 | 1531840 | 432 | - | PROTEIN:hypothetical protein |
| LCRIS_01522 | LCRIS_01522 | 1531837 | 1532271 | 435 | - | PROTEIN:response regulator of the lytr/algr family |
| LCRIS_01523 | LCRIS_01523 | 1532435 | 1532956 | 522 | - | PROTEIN:hypothetical protein |
| LCRIS_01524 | LCRIS_01524 | 1533163 | 1534092 | 930 | - | PROTEIN:hypothetical protein |
| LCRIS_01525 | thrS | 1534222 | 1536159 | 1938 | - | PROTEIN:threonyl-tRNA synthetase |
| LCRIS_01526 | dnaI | 1536444 | 1537352 | 909 | - | PROTEIN:primosomal protein dnai |
| LCRIS_01527 | dnaB2 | 1537372 | 1538706 | 1335 | - | PROTEIN:replicative DNA helicase loader dnab |
| LCRIS_01528 | nrdR | 1538709 | 1539176 | 468 | - | PROTEIN:transcriptional repressor nrdr |
| LCRIS_01529 | coaE | 1539179 | 1539781 | 603 | - | PROTEIN:dephospho-CoA kinase |
| LCRIS_01530 | fpg | 1539781 | 1540608 | 828 | - | PROTEIN:formamidopyrimidine-DNA glycosylase |
| LCRIS_01531 | polA | 1540617 | 1543280 | 2664 | - | PROTEIN:DNA polymerase i |
| LCRIS_01532 | LCRIS_01532 | 1543487 | 1544368 | 882 | - | PROTEIN:membrane protease subunit, stomatin/prohib |
| LCRIS_01533 | LCRIS_01533 | 1544389 | 1544697 | 309 | - | PROTEIN:DNA-damage-inducible protein j |
| LCRIS_01534 | LCRIS_01534 | 1544811 | 1545791 | 981 | - | PROTEIN:bacteriocin helveticin j |
| LCRIS_01535 | LCRIS_01535 | 1545926 | 1546240 | 315 | + | PROTEIN:hypothetical protein |
| LCRIS_01536 | LCRIS_01536 | 1546343 | 1546831 | 489 | + | PROTEIN:hypothetical protein |
| LCRIS_01537 | LCRIS_01537 | 1546832 | 1547425 | 594 | + | PROTEIN:membrane alanine aminopeptidase |
| LCRIS_01538 | LCRIS_01538 | 1547539 | 1548516 | 978 | + | PROTEIN:surface protein |
| LCRIS_01539 | LCRIS_01539 | 1548624 | 1549958 | 1335 | + | PROTEIN:major facilitator superfamily, na-driven e |
| LCRIS_01540 | murC | 1550015 | 1551328 | 1314 | - | PROTEIN:UDP-N-acetylmuramate--l-alanine ligase |
| LCRIS_01541 | LCRIS_01541 | 1551616 | 1552263 | 648 | - | PROTEIN:phenylalanyl-tRNA synthetase |
| LCRIS_01542 | trx | 1552283 | 1552603 | 321 | - | PROTEIN:thioredoxin |
| LCRIS_01543 | LCRIS_01543 | 1552673 | 1552831 | 159 | - | PROTEIN:hypothetical protein |
| LCRIS_01544 | LCRIS_01544 | 1552949 | 1553176 | 228 | - | PROTEIN:hypothetical protein |
| LCRIS_01545 | trmB | 1553319 | 1553975 | 657 | - | PROTEIN:tRNA (guanine-n(7)-)-methyltransferase |
| LCRIS_01546 | LCRIS_01546 | 1553985 | 1555202 | 1218 | - | PROTEIN:ABC-type exoprotein transport system, perm |
| LCRIS_01547 | ecsA | 1555189 | 1555932 | 744 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01548 | hit | 1556023 | 1556454 | 432 | + | PROTEIN:histidine triad (hit) family protein |
| LCRIS_01549 | LCRIS_01549 | 1556464 | 1556820 | 357 | + | PROTEIN:tropomyosin |
| LCRIS_01550 | prsA | 1556939 | 1557835 | 897 | + | PROTEIN:parvulin-like peptidyl-prolyl isomerase |
| LCRIS_01551 | LCRIS_01551 | 1557853 | 1559550 | 1698 | - | PROTEIN:transposase |
| LCRIS_01552 | eda | 1559807 | 1560214 | 408 | - | PROTEIN:2-dehydro-3-deoxy-phosphogluconate aldolas |
| LCRIS_01553 | LCRIS_01553 | 1560322 | 1561800 | 1479 | - | PROTEIN:hypothetical protein |
| LCRIS_01554 | cbf | 1562035 | 1563012 | 978 | - | PROTEIN:cmp-binding factor |
| LCRIS_01555 | LCRIS_01555 | 1563005 | 1565500 | 2496 | - | PROTEIN:DNA repair atpase |
| LCRIS_01556 | LCRIS_01556 | 1565481 | 1566698 | 1218 | - | PROTEIN:phosphoesterase |
| LCRIS_01557 | LCRIS_01557 | 1566707 | 1567057 | 351 | - | PROTEIN:hypothetical protein |
| LCRIS_01558 | pbp3 | 1567074 | 1569131 | 2058 | - | PROTEIN:penicillin-binding protein |
| LCRIS_01559 | rluD3 | 1569219 | 1570076 | 858 | + | PROTEIN:pseudouridine synthase |
| LCRIS_01560 | ppsA | 1570131 | 1572503 | 2373 | - | PROTEIN:phosphoenolpyruvate synthase |
| LCRIS_01561 | LCRIS_01561 | 1572520 | 1573125 | 606 | - | PROTEIN:phosphoglycerate mutase |
| LCRIS_01562 | glpF | 1573557 | 1574309 | 753 | + | PROTEIN:glycerol uptake facilitator protein |
| LCRIS_01563 | celB5 | 1574361 | 1575701 | 1341 | - | PROTEIN:PTS system cellobiose-specific iic compone |
| LCRIS_01564 | bglA5 | 1575980 | 1577407 | 1428 | + | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_01565 | LCRIS_01565 | 1577473 | 1577802 | 330 | - | PROTEIN:hypothetical protein |
| LCRIS_01566 | citG | 1577892 | 1578710 | 819 | - | PROTEIN:triphosphoribosyl-dephospho-CoA synthase |
| LCRIS_01567 | LCRIS_01567 | 1578721 | 1579518 | 798 | - | PROTEIN:alpha/beta superfamily hydrolase |
| LCRIS_01568 | LCRIS_01568 | 1579520 | 1580182 | 663 | - | PROTEIN:phosphoribosyl-dephospho-CoA transferase |
| LCRIS_01569 | mdcD | 1580170 | 1581837 | 1668 | - | PROTEIN:malonate decarboxylase, beta subunit |
| LCRIS_01570 | mdcC | 1581827 | 1582126 | 300 | - | PROTEIN:malonate decarboxylase, delta subunit |
| LCRIS_01571 | mdcA | 1582145 | 1583788 | 1644 | - | PROTEIN:malonate decarboxylase, alpha subunit |
| LCRIS_01572 | fabD | 1583788 | 1584696 | 909 | - | PROTEIN:acyl transferase region |
| LCRIS_01573 | LCRIS_01573 | 1584808 | 1585695 | 888 | + | PROTEIN:similar to transcriptional regulator |
| LCRIS_01574 | LCRIS_01574 | 1585780 | 1586889 | 1110 | + | PROTEIN:integral hypothetical protein |
| LCRIS_01575 | LCRIS_01575 | 1586969 | 1588273 | 1305 | - | PROTEIN:PTS system iic component |
| LCRIS_01576 | LCRIS_01576 | 1588443 | 1589210 | 768 | + | PROTEIN:transcriptional regulator |
| LCRIS_01577 | LCRIS_01577 | 1589242 | 1589523 | 282 | - | PROTEIN:small conserved hypothetical protein |
| LCRIS_01578 | LCRIS_01578 | 1589543 | 1590550 | 1008 | - | PROTEIN:transcriptional regulator-type |
| LCRIS_01579 | nagZ | 1590740 | 1592443 | 1704 | + | PROTEIN:beta-hexosamidase a, glycoside hydrolase f |
| LCRIS_01580 | LCRIS_01580 | 1592567 | 1593217 | 651 | + | PROTEIN:hypothetical protein |
| LCRIS_01581 | LCRIS_01581 | 1593307 | 1593639 | 333 | + | PROTEIN:hypothetical protein |
| LCRIS_01582 | LCRIS_01582 | 1593786 | 1595324 | 1539 | - | PROTEIN:glycoside hydrolase |
| LCRIS_01583 | LCRIS_01583 | 1595346 | 1596566 | 1221 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01584 | rbsR | 1596742 | 1597701 | 960 | + | PROTEIN:ribose operon repressor |
| LCRIS_01585 | rpiA3 | 1597768 | 1598406 | 639 | - | PROTEIN:ribose-5-phosphate isomerase a |
| LCRIS_01586 | rpe2 | 1598457 | 1599134 | 678 | - | PROTEIN:ribulose-phosphate 3-epimerase |
| LCRIS_01587 | LCRIS_01587 | 1599293 | 1600060 | 768 | - | PROTEIN:cellobiose phosphotransferase |
| LCRIS_01588 | LCRIS_01588 | 1600112 | 1601020 | 909 | - | PROTEIN:sugar hydrolase |
| LCRIS_01589 | bglF2 | 1601032 | 1603236 | 2205 | - | PROTEIN:PTS system beta-glucoside-specific iiABC c |
| LCRIS_01590 | bglG2 | 1603229 | 1604083 | 855 | - | PROTEIN:beta-glucoside transcriptional antitermina |
| LCRIS_01591 | LCRIS_01591 | 1604269 | 1605567 | 1299 | - | PROTEIN:beta-glucosidase |
| LCRIS_01592 | LCRIS_01592 | 1605594 | 1607048 | 1455 | - | PROTEIN:l-fucose isomerase |
| LCRIS_01593 | LCRIS_01593 | 1607159 | 1608175 | 1017 | - | PROTEIN:ribose operon repressor |
| LCRIS_01594 | LCRIS_01594 | 1608388 | 1609011 | 624 | + | PROTEIN:transposase |
| LCRIS_01595 | LCRIS_01595 | 1609133 | 1610164 | 1032 | + | PROTEIN:transposase |
| LCRIS_01596 | LCRIS_01596 | 1610094 | 1610891 | 798 | - | PROTEIN:degv family protein |
| LCRIS_01597 | LCRIS_01597 | 1611077 | 1611562 | 486 | + | PROTEIN:hypothetical protein |
| LCRIS_01598 | LCRIS_01598 | 1612051 | 1613085 | 1035 | + | PROTEIN:transposase |
| LCRIS_01599 | LCRIS_01599 | 1613100 | 1615298 | 2199 | - | PROTEIN:hypothetical protein |
| LCRIS_01600 | celB6 | 1615298 | 1616590 | 1293 | - | PROTEIN:PTS system lactose/cellobiose-specific iic |
| LCRIS_01601 | LCRIS_01601 | 1616709 | 1617683 | 975 | + | PROTEIN:arac-type DNA-binding domain-containing pr |
| LCRIS_01602 | LCRIS_01602 | 1617696 | 1617962 | 267 | - | PROTEIN:small conserved hypothetical protein |
| LCRIS_01603 | LCRIS_01603 | 1617981 | 1618901 | 921 | - | PROTEIN:rok family protein |
| LCRIS_01604 | LCRIS_01604 | 1618894 | 1619628 | 735 | - | PROTEIN:hypothetical protein |
| LCRIS_01605 | LCRIS_01605 | 1619632 | 1619787 | 156 | - | PROTEIN:hypothetical protein |
| LCRIS_01606 | LCRIS_01606 | 1619799 | 1620995 | 1197 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01607 | LCRIS_01607 | 1620992 | 1623754 | 2763 | - | PROTEIN:alfa-l-rhamnosidase |
| LCRIS_01608 | LCRIS_01608 | 1623870 | 1624961 | 1092 | + | PROTEIN:arac-type DNA-binding domain-containing pr |
| LCRIS_01609 | LCRIS_01609 | 1625008 | 1625832 | 825 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01610 | LCRIS_01610 | 1625866 | 1626057 | 192 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01611 | LCRIS_01611 | 1626175 | 1626378 | 204 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01612 | LCRIS_01612 | 1626516 | 1627157 | 642 | - | PROTEIN:hypothetical protein |
| LCRIS_01613 | LCRIS_01613 | 1627243 | 1627566 | 324 | - | PROTEIN:hypothetical protein |
| LCRIS_01614 | LCRIS_01614 | 1627600 | 1628223 | 624 | - | PROTEIN:hypothetical protein |
| LCRIS_01615 | LCRIS_01615 | 1628445 | 1629224 | 780 | - | PROTEIN:response regulator bacteriocinproduction-r |
| LCRIS_01616 | LCRIS_01616 | 1629226 | 1629918 | 693 | - | PROTEIN:sensor histidine kinase |
| LCRIS_01617 | LCRIS_01617 | 1630553 | 1630840 | 288 | - | PROTEIN:hypothetical protein |
| LCRIS_01618 | LCRIS_01618 | 1631039 | 1631362 | 324 | - | PROTEIN:hypothetical protein |
| LCRIS_01619 | LCRIS_01619 | 1631433 | 1631981 | 549 | - | PROTEIN:1,4-dihydroxy-2-naphthoate octaprenyltrans |
| LCRIS_01620 | LCRIS_01620 | 1631984 | 1632343 | 360 | - | PROTEIN:1,4-dihydroxy-2-naphthoate octaprenyltrans |
| LCRIS_01621 | LCRIS_01621 | 1632622 | 1633158 | 537 | - | PROTEIN:mutt/nudix family protein |
| LCRIS_01622 | fbaA | 1633222 | 1634133 | 912 | - | PROTEIN:fructose-bisphosphate aldolase |
| LCRIS_01623 | argS | 1634244 | 1635929 | 1686 | - | PROTEIN:arginyl-tRNA synthetase |
| LCRIS_01624 | LCRIS_01624 | 1636293 | 1636760 | 468 | - | PROTEIN:erfk/ybis/ycfs/ynhg superfamily |
| LCRIS_01625 | LCRIS_01625 | 1637104 | 1637952 | 849 | - | PROTEIN:sugar kinase |
| LCRIS_01626 | dacA | 1637955 | 1639256 | 1302 | - | PROTEIN:D-alanyl-D-alanine carboxypeptidase |
| LCRIS_02088 | tRNA-Leu | 1639345 | 1639431 | 87 | + | RNA:Leu tRNA |
| LCRIS_01627 | LCRIS_01627 | 1639566 | 1639886 | 321 | + | PROTEIN:phage shock protein c, pspc |
| LCRIS_01628 | LCRIS_01628 | 1640274 | 1641920 | 1647 | - | PROTEIN:polysaccharide transporter |
| LCRIS_01629 | leuS | 1642014 | 1644428 | 2415 | - | PROTEIN:leucyl-tRNA synthetase |
| LCRIS_01630 | LCRIS_01630 | 1644718 | 1645383 | 666 | + | PROTEIN:phospholipid phosphatase |
| LCRIS_01631 | LCRIS_01631 | 1645426 | 1646349 | 924 | - | PROTEIN:transcriptional regulator, lysr family |
| LCRIS_01632 | LCRIS_01632 | 1646410 | 1646937 | 528 | - | PROTEIN:hypothetical protein |
| LCRIS_01633 | ndh | 1647119 | 1648339 | 1221 | - | PROTEIN:pyridine nucleotide-disulphide oxidoreduct |
| LCRIS_01634 | menA | 1648354 | 1649253 | 900 | - | PROTEIN:prenyltransferase, ubia family |
| LCRIS_01635 | LCRIS_01635 | 1649313 | 1650302 | 990 | + | PROTEIN:polyprenyl synthetase |
| LCRIS_01636 | ubiE | 1650299 | 1651000 | 702 | - | PROTEIN:ubiquinone/menaquinone biosynthesis methyl |
| LCRIS_01637 | LCRIS_01637 | 1651010 | 1652131 | 1122 | - | PROTEIN:ABC transporter, ATP-binding protein cydc |
| LCRIS_01638 | LCRIS_01638 | 1652221 | 1652757 | 537 | - | PROTEIN:ABC transporter, ATP-binding protein cydc |
| LCRIS_01639 | LCRIS_01639 | 1652747 | 1654465 | 1719 | - | PROTEIN:ABC transporter, ATP-binding protein cydd |
| LCRIS_01640 | cydB | 1654465 | 1655481 | 1017 | - | PROTEIN:cytochrome d ubiquinol oxidase, subunit ii |
| LCRIS_01641 | cydA | 1655483 | 1656913 | 1431 | - | PROTEIN:cytochrome d ubiquinol oxidase, subunit i |
| LCRIS_01642 | LCRIS_01642 | 1657240 | 1658085 | 846 | - | PROTEIN:transcriptional regulator, rpir family |
| LCRIS_01643 | LCRIS_01643 | 1658103 | 1659140 | 1038 | - | PROTEIN:hypothetical protein |
| LCRIS_01644 | murQ | 1659200 | 1660099 | 900 | - | PROTEIN:N-acetylmuramic acid 6-phosphate etherase |
| LCRIS_01645 | scrA3 | 1660123 | 1662093 | 1971 | - | PROTEIN:PTS system sucrose-specific iibc component |
| LCRIS_01646 | LCRIS_01646 | 1662321 | 1663781 | 1461 | - | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01647 | metK | 1663805 | 1665004 | 1200 | - | PROTEIN:s-adenosylmethionine synthetase |
| LCRIS_02094 | tRNA-Val | 1665162 | 1665234 | 73 | - | RNA:Val tRNA |
| LCRIS_02093 | tRNA-Glu | 1665237 | 1665311 | 75 | - | RNA:Glu tRNA |
| LCRIS_01648 | LCRIS_01648 | 1665463 | 1666350 | 888 | - | PROTEIN:ABC-type metal ion transport system, subst |
| LCRIS_02092 | tRNA-Ser | 1666439 | 1666529 | 91 | - | RNA:Ser tRNA |
| LCRIS_02091 | tRNA-Asn | 1666540 | 1666612 | 73 | - | RNA:Asn tRNA |
| LCRIS_02026 | 5srRNA | 1666625 | 1666740 | 116 | - | RNA:5S ribosomal RNA |
| LCRIS_02030 | 23srRNA | 1666813 | 1669718 | 2906 | - | RNA:23S ribosomal RNA |
| LCRIS_01649 | LCRIS_01649 | 1666956 | 1667273 | 318 | + | PROTEIN:cell wall-associated hydrolase |
| LCRIS_01650 | LCRIS_01650 | 1668694 | 1669164 | 471 | + | PROTEIN:hypothetical protein |
| LCRIS_02034 | 16srRNA | 1669931 | 1671482 | 1552 | - | RNA:16S ribosomal RNA |
| LCRIS_01651 | LCRIS_01651 | 1669999 | 1670352 | 354 | - | PROTEIN:hypothetical protein |
| LCRIS_01652 | LCRIS_01652 | 1670786 | 1671220 | 435 | + | PROTEIN:pg1 protein, homology to Homo sapiens |
| LCRIS_01653 | LCRIS_01653 | 1671280 | 1671405 | 126 | + | PROTEIN:hypothetical protein |
| LCRIS_01654 | LCRIS_01654 | 1672204 | 1682862 | 10659 | - | PROTEIN:mucus-binding protein |
| LCRIS_01655 | serS | 1683067 | 1684374 | 1308 | - | PROTEIN:seryl-tRNA synthetase |
| LCRIS_01656 | LCRIS_01656 | 1684605 | 1685156 | 552 | - | PROTEIN:ribosomal-protein-alanine N-acetyltransfer |
| LCRIS_01657 | LCRIS_01657 | 1685156 | 1685761 | 606 | - | PROTEIN:hypothetical protein |
| LCRIS_01658 | LCRIS_01658 | 1685919 | 1686713 | 795 | + | PROTEIN:esterase |
| LCRIS_01659 | LCRIS_01659 | 1686716 | 1687576 | 861 | + | PROTEIN:regulator |
| LCRIS_01660 | LCRIS_01660 | 1687604 | 1688107 | 504 | - | PROTEIN:deoxyribosyltransferase |
| LCRIS_01661 | gabD | 1688274 | 1689656 | 1383 | + | PROTEIN:aldehyde dehydrogenase |
| LCRIS_01662 | LCRIS_01662 | 1689931 | 1691157 | 1227 | + | PROTEIN:transposase |
| LCRIS_01663 | LCRIS_01663 | 1691392 | 1692408 | 1017 | - | PROTEIN:hypothetical protein |
| LCRIS_01664 | LCRIS_01664 | 1692522 | 1696280 | 3759 | - | PROTEIN:hypothetical protein |
| LCRIS_01665 | LCRIS_01665 | 1696285 | 1696860 | 576 | - | PROTEIN:transcriptional regulator |
| LCRIS_01666 | penP | 1696993 | 1697937 | 945 | - | PROTEIN:beta-lactamase |
| LCRIS_01667 | LCRIS_01667 | 1697953 | 1698870 | 918 | - | PROTEIN:Mg+2 and co2 transporter |
| LCRIS_01668 | LCRIS_01668 | 1698989 | 1700290 | 1302 | - | PROTEIN:glycerol-3-phosphate ABC transporter |
| LCRIS_01669 | LCRIS_01669 | 1700344 | 1701639 | 1296 | - | PROTEIN:hydrolase of the metallo-beta-lactamase su |
| LCRIS_01670 | LCRIS_01670 | 1701623 | 1702447 | 825 | - | PROTEIN:ABC transporter, permease protein |
| LCRIS_01671 | LCRIS_01671 | 1702451 | 1703317 | 867 | - | PROTEIN:sugar ABC transporter, permease protein |
| LCRIS_01672 | LCRIS_01672 | 1703317 | 1704405 | 1089 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01673 | LCRIS_01673 | 1704662 | 1706089 | 1428 | - | PROTEIN:dipeptidase |
| LCRIS_01674 | LCRIS_01674 | 1706146 | 1707000 | 855 | - | PROTEIN:DNA-entry nuclease |
| LCRIS_01675 | dnaQ2 | 1707010 | 1707966 | 957 | - | PROTEIN:DNA polymerase iii |
| LCRIS_01676 | LCRIS_01676 | 1707982 | 1708803 | 822 | - | PROTEIN:nad-dependent protein deacetylase, sir2 fa |
| LCRIS_01677 | LCRIS_01677 | 1708805 | 1709452 | 648 | - | PROTEIN:hypothetical protein |
| LCRIS_01678 | LCRIS_01678 | 1709452 | 1709901 | 450 | - | PROTEIN:hypothetical protein |
| LCRIS_01679 | pnuC | 1710005 | 1710745 | 741 | - | PROTEIN:nicotinamide mononucleotide transporter |
| LCRIS_01680 | bglA6 | 1710984 | 1712459 | 1476 | - | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_01681 | bglF3 | 1712470 | 1714377 | 1908 | - | PROTEIN:PTS system beta-glucoside-specific iiABC c |
| LCRIS_01682 | bglG3 | 1714374 | 1715228 | 855 | - | PROTEIN:transcriptional antiterminator |
| LCRIS_01683 | LCRIS_01683 | 1715790 | 1716014 | 225 | + | PROTEIN:hypothetical protein |
| LCRIS_01684 | LCRIS_01684 | 1716066 | 1716806 | 741 | - | PROTEIN:flavoprotein |
| LCRIS_01685 | LCRIS_01685 | 1716958 | 1718562 | 1605 | - | PROTEIN:ABC transporter, ATP-binding/permease prot |
| LCRIS_01686 | pip | 1718684 | 1719601 | 918 | + | PROTEIN:prolyl aminopeptidase |
| LCRIS_01687 | LCRIS_01687 | 1719659 | 1720378 | 720 | + | PROTEIN:response regulator |
| LCRIS_01688 | LCRIS_01688 | 1720365 | 1721699 | 1335 | + | PROTEIN:sensor protein |
| LCRIS_01689 | LCRIS_01689 | 1721818 | 1722366 | 549 | + | PROTEIN:hypothetical protein |
| LCRIS_01690 | LCRIS_01690 | 1722384 | 1722995 | 612 | + | PROTEIN:lipoprotein |
| LCRIS_01691 | LCRIS_01691 | 1723057 | 1723581 | 525 | - | PROTEIN:acetyltransferase, gnat family protein |
| LCRIS_01692 | LCRIS_01692 | 1723581 | 1724036 | 456 | - | PROTEIN:transcriptional regulator, marr family pro |
| LCRIS_01693 | LCRIS_01693 | 1724127 | 1725773 | 1647 | - | PROTEIN:oligopeptide ABC transporter, oligopeptide |
| LCRIS_01694 | LCRIS_01694 | 1726074 | 1726469 | 396 | + | PROTEIN:hypothetical protein |
| LCRIS_01695 | LCRIS_01695 | 1726596 | 1727183 | 588 | + | PROTEIN:hypothetical protein |
| LCRIS_01696 | efp2 | 1727230 | 1727805 | 576 | - | PROTEIN:elongation factor p |
| LCRIS_01697 | coaA | 1727923 | 1728837 | 915 | - | PROTEIN:pantothenate kinase |
| LCRIS_01698 | LCRIS_01698 | 1728897 | 1729454 | 558 | + | PROTEIN:arylesterase |
| LCRIS_01699 | LCRIS_01699 | 1729451 | 1729900 | 450 | - | PROTEIN:acetyltransferase, gnat family |
| LCRIS_01700 | LCRIS_01700 | 1729902 | 1730717 | 816 | - | PROTEIN:amino acid ABC transporter, amino acid-bin |
| LCRIS_01701 | LCRIS_01701 | 1730721 | 1731341 | 621 | - | PROTEIN:amino acid ABC transporter, ATP-binding pr |
| LCRIS_01702 | LCRIS_01702 | 1731355 | 1732002 | 648 | - | PROTEIN:amino acid ABC transporter, permease prote |
| LCRIS_01703 | uvrD | 1732436 | 1734742 | 2307 | - | PROTEIN:ATP-dependent DNA helicase |
| LCRIS_01704 | LCRIS_01704 | 1734895 | 1735587 | 693 | + | PROTEIN:metalloprotease |
| LCRIS_01705 | gpm3 | 1735648 | 1736298 | 651 | + | PROTEIN:phosphoglycerate mutase |
| LCRIS_01706 | LCRIS_01706 | 1736441 | 1737712 | 1272 | - | PROTEIN:hypothetical protein |
| LCRIS_01707 | LCRIS_01707 | 1737853 | 1738539 | 687 | + | PROTEIN:hypothetical protein |
| LCRIS_01708 | LCRIS_01708 | 1738719 | 1741370 | 2652 | + | PROTEIN:cation-transporting atpase, p-type |
| LCRIS_01709 | manX3 | 1741420 | 1741818 | 399 | - | PROTEIN:PTS system iia component |
| LCRIS_01710 | LCRIS_01710 | 1741874 | 1742167 | 294 | - | PROTEIN:hypothetical protein |
| LCRIS_01711 | ansA | 1742454 | 1743446 | 993 | - | PROTEIN:l-asparaginase |
| LCRIS_01712 | LCRIS_01712 | 1743458 | 1744231 | 774 | - | PROTEIN:transcriptional regulator |
| LCRIS_01713 | glvA | 1744363 | 1745700 | 1338 | - | PROTEIN:maltose-6'-phosphate glucosidase |
| LCRIS_01714 | LCRIS_01714 | 1746522 | 1747355 | 834 | + | PROTEIN:hypothetical protein |
| LCRIS_01715 | LCRIS_01715 | 1747812 | 1748354 | 543 | + | PROTEIN:transcriptional regulator |
| LCRIS_01716 | hpt2 | 1748380 | 1748913 | 534 | - | PROTEIN:hypoxanthine phosphoribosyltransferase |
| LCRIS_01717 | LCRIS_01717 | 1749152 | 1749718 | 567 | - | PROTEIN:acetyltransferase |
| LCRIS_01718 | LCRIS_01718 | 1749736 | 1751883 | 2148 | - | PROTEIN:hypothetical protein |
| LCRIS_01719 | LCRIS_01719 | 1752121 | 1752273 | 153 | - | PROTEIN:hypothetical protein |
| LCRIS_01720 | LCRIS_01720 | 1752348 | 1752638 | 291 | - | PROTEIN:polysaccharide biosynthesis protein |
| LCRIS_01721 | LCRIS_01721 | 1752656 | 1753240 | 585 | + | PROTEIN:hypothetical protein |
| LCRIS_01722 | LCRIS_01722 | 1753308 | 1755782 | 2475 | - | PROTEIN:5'-nucleotidase/2',3'-cyclic phosphodieste |
| LCRIS_01723 | LCRIS_01723 | 1756098 | 1756463 | 366 | - | PROTEIN:transposase |
| LCRIS_01724 | LCRIS_01724 | 1756705 | 1757034 | 330 | + | PROTEIN:transposase |
| LCRIS_01725 | LCRIS_01725 | 1757118 | 1757609 | 492 | + | PROTEIN:transposase |
| LCRIS_01726 | LCRIS_01726 | 1757684 | 1757866 | 183 | + | PROTEIN:isef1, transposase |
| LCRIS_01727 | LCRIS_01727 | 1758141 | 1759286 | 1146 | - | PROTEIN:transcriptional regulator |
| LCRIS_01728 | LCRIS_01728 | 1759270 | 1760001 | 732 | - | PROTEIN:hypothetical protein |
| LCRIS_01729 | LCRIS_01729 | 1760176 | 1760400 | 225 | + | PROTEIN:hypothetical protein |
| LCRIS_01730 | LCRIS_01730 | 1760411 | 1760632 | 222 | + | PROTEIN:hypothetical protein |
| LCRIS_01731 | LCRIS_01731 | 1760711 | 1760914 | 204 | + | PROTEIN:hypothetical protein |
| LCRIS_01732 | LCRIS_01732 | 1761259 | 1762485 | 1227 | - | PROTEIN:transposase |
| LCRIS_01733 | rfbD | 1762749 | 1763735 | 987 | - | PROTEIN:dtdp-4-dehydrorhamnose reductase |
| LCRIS_01734 | rfbC | 1763756 | 1764364 | 609 | - | PROTEIN:dtdp-4-dehydrorhamnose 3,5-epimerase |
| LCRIS_01735 | rfbA | 1764390 | 1765274 | 885 | - | PROTEIN:glucose-1-phosphate thymidylyltransferase |
| LCRIS_01736 | rfbB | 1765277 | 1766314 | 1038 | - | PROTEIN:dtdp-glucose 4,6-dehydratase |
| LCRIS_01737 | LCRIS_01737 | 1766402 | 1767574 | 1173 | - | PROTEIN:transcriptional regulator |
| LCRIS_01738 | LCRIS_01738 | 1767674 | 1767979 | 306 | - | PROTEIN:hypothetical protein |
| LCRIS_01739 | LCRIS_01739 | 1767979 | 1768248 | 270 | - | PROTEIN:hypothetical protein |
| LCRIS_01740 | LCRIS_01740 | 1768573 | 1768968 | 396 | - | PROTEIN:hypothetical protein |
| LCRIS_01741 | sacB | 1769344 | 1771323 | 1980 | - | PROTEIN:levansucrase |
| LCRIS_01742 | LCRIS_01742 | 1771580 | 1772041 | 462 | - | PROTEIN:hypothetical protein |
| LCRIS_01743 | LCRIS_01743 | 1772202 | 1773380 | 1179 | - | PROTEIN:transposase |
| LCRIS_01744 | LCRIS_01744 | 1773499 | 1773876 | 378 | - | PROTEIN:hypothetical protein |
| LCRIS_01745 | LCRIS_01745 | 1774343 | 1774708 | 366 | + | PROTEIN:hypothetical protein |
| LCRIS_01746 | LCRIS_01746 | 1775153 | 1776385 | 1233 | + | PROTEIN:transposase |
| LCRIS_01747 | LCRIS_01747 | 1776827 | 1777975 | 1149 | + | PROTEIN:transposase |
| LCRIS_01748 | LCRIS_01748 | 1778215 | 1779699 | 1485 | - | PROTEIN:hypothetical protein |
| LCRIS_01749 | LCRIS_01749 | 1779711 | 1780718 | 1008 | - | PROTEIN:hypothetical protein |
| LCRIS_01750 | LCRIS_01750 | 1781769 | 1783103 | 1335 | - | PROTEIN:hypothetical protein |
| LCRIS_01751 | LCRIS_01751 | 1783164 | 1784591 | 1428 | - | PROTEIN:polysaccharide transporter, pst family |
| LCRIS_01752 | glf | 1784599 | 1785708 | 1110 | - | PROTEIN:udp-galactopyranose mutase |
| LCRIS_01753 | LCRIS_01753 | 1785711 | 1786733 | 1023 | - | PROTEIN:hypothetical protein |
| LCRIS_01754 | LCRIS_01754 | 1786714 | 1788108 | 1395 | - | PROTEIN:hypothetical protein |
| LCRIS_01755 | LCRIS_01755 | 1788128 | 1788931 | 804 | - | PROTEIN:glycosyl transferase, group 2 |
| LCRIS_01756 | LCRIS_01756 | 1788943 | 1789695 | 753 | - | PROTEIN:exopolysaccharide biosynthesis protein |
| LCRIS_01757 | LCRIS_01757 | 1789723 | 1790520 | 798 | - | PROTEIN:lipopolysaccharide biosynthesis glycosyltr |
| LCRIS_01758 | LCRIS_01758 | 1790544 | 1791197 | 654 | - | PROTEIN:lipopolysaccharide synthesis sugar transfe |
| LCRIS_01759 | LCRIS_01759 | 1791300 | 1792070 | 771 | - | PROTEIN:exopolysaccharide biosynthesis protein |
| LCRIS_01760 | LCRIS_01760 | 1792070 | 1792855 | 786 | - | PROTEIN:exopolysaccharide biosynthesis protein |
| LCRIS_01761 | LCRIS_01761 | 1792870 | 1793748 | 879 | - | PROTEIN:exopolysaccharide biosynthesis protein |
| LCRIS_01762 | epsA | 1793766 | 1794818 | 1053 | - | PROTEIN:exopolysaccharide biosynthesis protein |
| LCRIS_01763 | LCRIS_01763 | 1794884 | 1795918 | 1035 | - | PROTEIN:transposase |
| LCRIS_01764 | yoeB | 1796173 | 1796445 | 273 | - | PROTEIN:addiction module toxin, txe/yoeb family |
| LCRIS_01765 | LCRIS_01765 | 1796445 | 1796705 | 261 | - | PROTEIN:addiction module antitoxin, relb/dinj fami |
| LCRIS_01766 | hflX | 1797077 | 1798342 | 1266 | + | PROTEIN:GTP-binding protein |
| LCRIS_01767 | LCRIS_01767 | 1798428 | 1799387 | 960 | + | PROTEIN:hypothetical protein |
| LCRIS_01768 | LCRIS_01768 | 1799899 | 1800120 | 222 | - | PROTEIN:hypothetical protein |
| LCRIS_01769 | LCRIS_01769 | 1800452 | 1801342 | 891 | - | PROTEIN:cell wall-associated hydrolase |
| LCRIS_01770 | LCRIS_01770 | 1801512 | 1802255 | 744 | - | PROTEIN:cell wall-associated hydrolase |
| LCRIS_01771 | LCRIS_01771 | 1802525 | 1803076 | 552 | - | PROTEIN:glycosidase |
| LCRIS_01772 | gmk2 | 1803249 | 1803791 | 543 | - | PROTEIN:guanylate kinase |
| LCRIS_01773 | LCRIS_01773 | 1803887 | 1804189 | 303 | + | PROTEIN:hypothetical protein |
| LCRIS_01774 | LCRIS_01774 | 1804275 | 1805783 | 1509 | + | PROTEIN:hypothetical protein |
| LCRIS_01775 | LCRIS_01775 | 1805837 | 1806460 | 624 | - | PROTEIN:hypothetical protein |
| LCRIS_01776 | LCRIS_01776 | 1806649 | 1807800 | 1152 | - | PROTEIN:transposase |
| LCRIS_01777 | nrdI | 1808093 | 1808539 | 447 | + | PROTEIN:ribonucleotide reductase |
| LCRIS_01778 | nrdF | 1808542 | 1809477 | 936 | + | PROTEIN:ribonucleotide reductase |
| LCRIS_01779 | LCRIS_01779 | 1809517 | 1810047 | 531 | - | PROTEIN:hypothetical protein |
| LCRIS_01780 | LCRIS_01780 | 1810163 | 1810876 | 714 | + | PROTEIN:yvpb-like protein |
| LCRIS_01781 | LCRIS_01781 | 1811072 | 1811557 | 486 | - | PROTEIN:hypothetical protein |
| LCRIS_01782 | LCRIS_01782 | 1811560 | 1812330 | 771 | - | PROTEIN:hypothetical protein |
| LCRIS_01783 | LCRIS_01783 | 1812341 | 1813882 | 1542 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01784 | LCRIS_01784 | 1813900 | 1814277 | 378 | - | PROTEIN:hypothetical protein |
| LCRIS_01785 | LCRIS_01785 | 1814395 | 1814973 | 579 | + | PROTEIN:ribosomal-protein-serine acetyltransferase |
| LCRIS_01786 | LCRIS_01786 | 1815288 | 1815830 | 543 | + | PROTEIN:hypothetical protein |
| LCRIS_01787 | LCRIS_01787 | 1815827 | 1816516 | 690 | - | PROTEIN:transport protein |
| LCRIS_01788 | LCRIS_01788 | 1816581 | 1817819 | 1239 | - | PROTEIN:transcriptional regulator |
| LCRIS_01789 | pepF | 1817986 | 1819782 | 1797 | + | PROTEIN:oligoendopeptidase f |
| LCRIS_01790 | LCRIS_01790 | 1819856 | 1820035 | 180 | + | PROTEIN:hypothetical protein |
| LCRIS_01791 | LCRIS_01791 | 1820184 | 1820399 | 216 | + | PROTEIN:hypothetical protein |
| LCRIS_01792 | LCRIS_01792 | 1820403 | 1820810 | 408 | + | PROTEIN:death-on-curing family protein |
| LCRIS_01793 | LCRIS_01793 | 1820874 | 1821146 | 273 | - | PROTEIN:hypothetical protein |
| LCRIS_01794 | LCRIS_01794 | 1821224 | 1822021 | 798 | - | PROTEIN:abortive infection protein |
| LCRIS_01795 | LCRIS_01795 | 1822037 | 1822234 | 198 | - | PROTEIN:transcriptional regulator |
| LCRIS_01796 | LCRIS_01796 | 1822231 | 1822710 | 480 | - | PROTEIN:hypothetical protein |
| LCRIS_01797 | LCRIS_01797 | 1822964 | 1823122 | 159 | + | PROTEIN:hypothetical protein |
| LCRIS_01798 | LCRIS_01798 | 1823371 | 1824489 | 1119 | + | PROTEIN:methionine synthase, vitamin-b12 independe |
| LCRIS_01799 | luxS | 1824548 | 1825024 | 477 | + | PROTEIN:s-ribosylhomocysteine lyase |
| LCRIS_01800 | lctP2 | 1825110 | 1826624 | 1515 | - | PROTEIN:L-lactate permease |
| LCRIS_01801 | LCRIS_01801 | 1826881 | 1827675 | 795 | - | PROTEIN:ABC-type mn2 /zn2 transport system, permea |
| LCRIS_01802 | LCRIS_01802 | 1827675 | 1828322 | 648 | - | PROTEIN:ABC-type mn2 /zn2 transport system, atpase |
| LCRIS_01803 | LCRIS_01803 | 1828349 | 1829245 | 897 | - | PROTEIN:ABC-type metal ion transport system, subst |
| LCRIS_01804 | LCRIS_01804 | 1829498 | 1829785 | 288 | - | PROTEIN:transcriptional regulator, arsr family |
| LCRIS_01805 | fruA | 1829970 | 1831967 | 1998 | - | PROTEIN:PTS system fructose-specific iiABC compone |
| LCRIS_01806 | fruK | 1831988 | 1832902 | 915 | - | PROTEIN:1-phosphofructokinase |
| LCRIS_01807 | fruR | 1832904 | 1833659 | 756 | - | PROTEIN:fructose operon transcription regulator, d |
| LCRIS_01808 | LCRIS_01808 | 1833967 | 1835181 | 1215 | - | PROTEIN:ABC transporter, permease protein |
| LCRIS_01809 | LCRIS_01809 | 1835174 | 1835359 | 186 | - | PROTEIN:hypothetical protein |
| LCRIS_01810 | LCRIS_01810 | 1835402 | 1836070 | 669 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01811 | LCRIS_01811 | 1836486 | 1836587 | 102 | - | PROTEIN:hypothetical protein |
| LCRIS_01812 | LCRIS_01812 | 1836607 | 1837029 | 423 | - | PROTEIN:hypothetical protein |
| LCRIS_01813 | LCRIS_01813 | 1837063 | 1837257 | 195 | - | PROTEIN:hypothetical protein |
| LCRIS_01814 | LCRIS_01814 | 1837390 | 1837599 | 210 | - | PROTEIN:transcriptional regulator |
| LCRIS_01815 | LCRIS_01815 | 1837614 | 1838153 | 540 | - | PROTEIN:hypothetical protein |
| LCRIS_01816 | LCRIS_01816 | 1838158 | 1838298 | 141 | - | PROTEIN:hypothetical protein |
| LCRIS_01817 | LCRIS_01817 | 1838511 | 1838726 | 216 | - | PROTEIN:hypothetical protein |
| LCRIS_01818 | LCRIS_01818 | 1839008 | 1840042 | 1035 | - | PROTEIN:transposase |
| LCRIS_01819 | LCRIS_01819 | 1840443 | 1840667 | 225 | - | PROTEIN:hypothetical protein |
| LCRIS_01820 | LCRIS_01820 | 1841187 | 1841450 | 264 | - | PROTEIN:hypothetical protein |
| LCRIS_01821 | LCRIS_01821 | 1841462 | 1841866 | 405 | - | PROTEIN:hypothetical protein |
| LCRIS_01822 | LCRIS_01822 | 1842040 | 1842252 | 213 | - | PROTEIN:hypothetical protein |
| LCRIS_01823 | LCRIS_01823 | 1842276 | 1843076 | 801 | - | PROTEIN:lactacin f two-component system response r |
| LCRIS_01824 | LCRIS_01824 | 1843069 | 1844412 | 1344 | - | PROTEIN:lactacin f two-component system histidine |
| LCRIS_01825 | LCRIS_01825 | 1844706 | 1845917 | 1212 | - | PROTEIN:predicted metal-dependent membrane proteas |
| LCRIS_01826 | LCRIS_01826 | 1846073 | 1846240 | 168 | - | PROTEIN:hypothetical protein |
| LCRIS_01827 | LCRIS_01827 | 1846370 | 1847569 | 1200 | - | PROTEIN:predicted metal-dependent membrane proteas |
| LCRIS_01828 | LCRIS_01828 | 1847939 | 1848346 | 408 | - | PROTEIN:hypothetical protein |
| LCRIS_01829 | LCRIS_01829 | 1848478 | 1848663 | 186 | - | PROTEIN:hypothetical protein |
| LCRIS_01830 | malZ | 1848828 | 1851134 | 2307 | - | PROTEIN:alpha-glucosidase |
| LCRIS_01831 | glcD | 1851281 | 1852684 | 1404 | - | PROTEIN:glycolate oxidase |
| LCRIS_01832 | LCRIS_01832 | 1852892 | 1853077 | 186 | - | PROTEIN:hypothetical protein |
| LCRIS_01833 | LCRIS_01833 | 1853092 | 1853283 | 192 | - | PROTEIN:hypothetical protein |
| LCRIS_01834 | LCRIS_01834 | 1853359 | 1853583 | 225 | - | PROTEIN:hypothetical protein |
| LCRIS_01835 | LCRIS_01835 | 1853732 | 1854055 | 324 | - | PROTEIN:hypothetical protein |
| LCRIS_01836 | racD | 1854197 | 1854946 | 750 | - | PROTEIN:aspartate racemase |
| LCRIS_01837 | murE2 | 1854956 | 1856527 | 1572 | - | PROTEIN:UDP-N-acetylmuramoyl-l-alanyl-d-glutamate- |
| LCRIS_01838 | LCRIS_01838 | 1856580 | 1857734 | 1155 | - | PROTEIN:sensor protein |
| LCRIS_01839 | LCRIS_01839 | 1857736 | 1858422 | 687 | - | PROTEIN:response regulator |
| LCRIS_01840 | LCRIS_01840 | 1858694 | 1860514 | 1821 | - | PROTEIN:succinate dehydrogenase/fumarate reductase |
| LCRIS_01841 | crr2 | 1860526 | 1861032 | 507 | - | PROTEIN:PTS system glucose-specific iia component |
| LCRIS_01842 | bglA7 | 1861032 | 1862471 | 1440 | - | PROTEIN:6-phospho-beta-glucosidase |
| LCRIS_01843 | ascF | 1862487 | 1863884 | 1398 | - | PROTEIN:PTS system beta-glucosides-specific iiABC |
| LCRIS_01844 | bglG4 | 1863900 | 1864733 | 834 | - | PROTEIN:beta-glucoside operon antiterminator |
| LCRIS_01845 | LCRIS_01845 | 1864940 | 1866805 | 1866 | - | PROTEIN:ABC-transporter, atpase and permease compo |
| LCRIS_01846 | LCRIS_01846 | 1866815 | 1868539 | 1725 | - | PROTEIN:ABC-transporter, atpase and permease compo |
| LCRIS_01847 | LCRIS_01847 | 1868686 | 1869471 | 786 | - | PROTEIN:hypothetical protein |
| LCRIS_01848 | ychF | 1869480 | 1870580 | 1101 | - | PROTEIN:GTP-binding protein |
| LCRIS_01849 | LCRIS_01849 | 1870634 | 1870897 | 264 | - | PROTEIN:hypothetical protein |
| LCRIS_01850 | parB1 | 1870890 | 1871777 | 888 | - | PROTEIN:chromosome partitioning protein parb |
| LCRIS_01851 | parA | 1871755 | 1872534 | 780 | - | PROTEIN:chromosome partitioning protein para |
| LCRIS_01852 | parB2 | 1872553 | 1873389 | 837 | - | PROTEIN:chromosome partitioning protein |
| LCRIS_01853 | gidB | 1873407 | 1874129 | 723 | - | PROTEIN:ribosomal RNA small subunit methyltransfer |
| LCRIS_01854 | LCRIS_01854 | 1874291 | 1874827 | 537 | + | PROTEIN:membrane anchor connecting muts2 with cell |
| LCRIS_01855 | LCRIS_01855 | 1874946 | 1875122 | 177 | - | PROTEIN:hypothetical protein |
| LCRIS_01856 | apbE | 1875244 | 1876173 | 930 | - | PROTEIN:thiamine biosynthesis lipoprotein |
| LCRIS_01857 | LCRIS_01857 | 1876176 | 1876967 | 792 | - | PROTEIN:protein-tyrosine phosphatase |
| LCRIS_01858 | LCRIS_01858 | 1877059 | 1877607 | 549 | - | PROTEIN:oxidoreductase |
| LCRIS_01859 | LCRIS_01859 | 1877621 | 1878160 | 540 | - | PROTEIN:oxidoreductase |
| LCRIS_01860 | LCRIS_01860 | 1878313 | 1878936 | 624 | - | PROTEIN:transcriptional regulator |
| LCRIS_01861 | LCRIS_01861 | 1879067 | 1879309 | 243 | - | PROTEIN:hypothetical protein |
| LCRIS_01862 | pepD2 | 1879418 | 1880830 | 1413 | + | PROTEIN:dipeptidase |
| LCRIS_01863 | LCRIS_01863 | 1880863 | 1881048 | 186 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01864 | LCRIS_01864 | 1881332 | 1882624 | 1293 | - | PROTEIN:transposase |
| LCRIS_01865 | LCRIS_01865 | 1882655 | 1883269 | 615 | - | PROTEIN:resolvase, n terminal domain family protei |
| LCRIS_01866 | LCRIS_01866 | 1883346 | 1883498 | 153 | - | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01867 | LCRIS_01867 | 1883495 | 1883746 | 252 | - | PROTEIN:ABC transporter ATP-binding protein |
| LCRIS_01868 | LCRIS_01868 | 1883748 | 1884809 | 1062 | - | PROTEIN:ABC-type antimicrobial peptide transport s |
| LCRIS_01869 | LCRIS_01869 | 1884812 | 1885336 | 525 | - | PROTEIN:transcriptional regulator |
| LCRIS_01870 | gpm4 | 1885456 | 1886055 | 600 | - | PROTEIN:phosphoglycerate mutase |
| LCRIS_01871 | LCRIS_01871 | 1886185 | 1886940 | 756 | + | PROTEIN:hydrolase of alpha-beta family |
| LCRIS_01872 | LCRIS_01872 | 1886937 | 1887698 | 762 | - | PROTEIN:hypothetical protein |
| LCRIS_01873 | LCRIS_01873 | 1887757 | 1888401 | 645 | + | PROTEIN:hypothetical protein |
| LCRIS_01874 | LCRIS_01874 | 1888485 | 1889282 | 798 | - | PROTEIN:galactose-1-phosphate uridylyltransferase |
| LCRIS_01875 | LCRIS_01875 | 1889437 | 1890639 | 1203 | + | PROTEIN:secreted protein |
| LCRIS_01876 | LCRIS_01876 | 1890731 | 1890922 | 192 | + | PROTEIN:hypothetical protein |
| LCRIS_01877 | LCRIS_01877 | 1891100 | 1892401 | 1302 | + | PROTEIN:transposase |
| LCRIS_01878 | LCRIS_01878 | 1892595 | 1893368 | 774 | - | PROTEIN:hypothetical protein |
| LCRIS_01879 | LCRIS_01879 | 1893371 | 1893820 | 450 | - | PROTEIN:response regulator of the lytr/algr family |
| LCRIS_01880 | LCRIS_01880 | 1893961 | 1894179 | 219 | - | PROTEIN:small conserved hypothetical protein |
| LCRIS_01881 | LCRIS_01881 | 1894301 | 1895638 | 1338 | - | PROTEIN:di-/tripeptide transporter |
| LCRIS_01882 | pepN | 1895909 | 1898446 | 2538 | - | PROTEIN:aminopeptidase n |
| LCRIS_01883 | LCRIS_01883 | 1898660 | 1899022 | 363 | + | PROTEIN:aggregation promoting protein |
| LCRIS_01884 | LCRIS_01884 | 1899094 | 1900077 | 984 | - | PROTEIN:D-ala, D-ala ligase |
| LCRIS_01885 | mdtG2 | 1900230 | 1901477 | 1248 | + | PROTEIN:permease of the major facilitator superfam |
| LCRIS_01886 | LCRIS_01886 | 1901576 | 1902079 | 504 | - | PROTEIN:appr-1-p processing domain protein |
| LCRIS_01887 | LCRIS_01887 | 1902263 | 1903135 | 873 | - | PROTEIN:transposase |
| LCRIS_01888 | LCRIS_01888 | 1903132 | 1903440 | 309 | - | PROTEIN:transposase |
| LCRIS_01889 | LCRIS_01889 | 1903557 | 1904063 | 507 | - | PROTEIN:transcriptional regulator |
| LCRIS_01890 | malG | 1904173 | 1905030 | 858 | - | PROTEIN:maltose ABC transporter, permease protein |
| LCRIS_01891 | malF | 1905033 | 1906391 | 1359 | - | PROTEIN:maltose ABC transporter, permease protein |
| LCRIS_01892 | malE2 | 1906466 | 1907692 | 1227 | - | PROTEIN:maltose ABC transporter, maltose-binding p |
| LCRIS_01893 | malK | 1907807 | 1908913 | 1107 | - | PROTEIN:maltose ABC transporter, ATP-binding prote |
| LCRIS_01894 | pgmB | 1908935 | 1909597 | 663 | - | PROTEIN:beta-phosphoglucomutase |
| LCRIS_01895 | mapA | 1909590 | 1911854 | 2265 | - | PROTEIN:maltose phosphorylase |
| LCRIS_01896 | nplT | 1912026 | 1913747 | 1722 | - | PROTEIN:neopullulanase |
| LCRIS_01897 | malL | 1913751 | 1915412 | 1662 | - | PROTEIN:alpha,alpha-phosphotrehalase |
| LCRIS_01898 | LCRIS_01898 | 1915525 | 1915953 | 429 | - | PROTEIN:hypothetical protein |
| LCRIS_01899 | ackA3 | 1915954 | 1917132 | 1179 | - | PROTEIN:acetate kinase |
| LCRIS_01900 | LCRIS_01900 | 1917276 | 1918226 | 951 | - | PROTEIN:transcriptional regulator, laci family |
| LCRIS_01901 | LCRIS_01901 | 1918234 | 1919007 | 774 | - | PROTEIN:protein-tyrosine phosphatase |
| LCRIS_01902 | LCRIS_01902 | 1919153 | 1919866 | 714 | - | PROTEIN:hypothetical protein |
| LCRIS_01903 | LCRIS_01903 | 1919859 | 1921472 | 1614 | - | PROTEIN:ABC transporter, atpase and permease compo |
| LCRIS_01904 | LCRIS_01904 | 1921447 | 1921740 | 294 | - | PROTEIN:hypothetical protein |
| LCRIS_01905 | LCRIS_01905 | 1921896 | 1923074 | 1179 | - | PROTEIN:transposase |
| LCRIS_01906 | LCRIS_01906 | 1923286 | 1924077 | 792 | - | PROTEIN:transcriptional regulator |
| LCRIS_01907 | glpK | 1924122 | 1925624 | 1503 | - | PROTEIN:glycerol kinase |
| LCRIS_01908 | thiD | 1925749 | 1926564 | 816 | + | PROTEIN:phosphomethylpyrimidine kinase |
| LCRIS_01909 | LCRIS_01909 | 1926893 | 1927435 | 543 | - | PROTEIN:hypothetical protein |
| LCRIS_01910 | LCRIS_01910 | 1927468 | 1927701 | 234 | - | PROTEIN:hypothetical protein |
| LCRIS_01911 | LCRIS_01911 | 1927799 | 1928632 | 834 | - | PROTEIN:transcriptional regulator, xre family |
| LCRIS_01912 | LCRIS_01912 | 1928734 | 1928952 | 219 | - | PROTEIN:addiction module toxin, txe/yoeb family |
| LCRIS_01913 | LCRIS_01913 | 1928988 | 1929263 | 276 | - | PROTEIN:prevent-host-death family protein |
| LCRIS_01914 | LCRIS_01914 | 1929700 | 1930110 | 411 | + | PROTEIN:hypothetical protein |
| LCRIS_01915 | LCRIS_01915 | 1930116 | 1930409 | 294 | + | PROTEIN:pare-domain protein |
| LCRIS_01916 | LCRIS_01916 | 1930479 | 1931165 | 687 | + | PROTEIN:cobyric acid synthase |
| LCRIS_01917 | LCRIS_01917 | 1931210 | 1931695 | 486 | - | PROTEIN:nlp-p60 family secreted protein |
| LCRIS_01918 | LCRIS_01918 | 1931890 | 1932240 | 351 | + | PROTEIN:hypothetical protein |
| LCRIS_01919 | LCRIS_01919 | 1932361 | 1933239 | 879 | + | PROTEIN:transcriptional regulator |
| LCRIS_01920 | LCRIS_01920 | 1933853 | 1934134 | 282 | + | PROTEIN:hypothetical protein |
| LCRIS_01921 | LCRIS_01921 | 1934153 | 1935082 | 930 | - | PROTEIN:permease of the drug/metabolite transporte |
| LCRIS_01922 | LCRIS_01922 | 1935160 | 1936080 | 921 | - | PROTEIN:permease of the drug/metabolite transporte |
| LCRIS_01923 | LCRIS_01923 | 1936163 | 1936828 | 666 | - | PROTEIN:transport protein |
| LCRIS_01924 | LCRIS_01924 | 1936894 | 1937538 | 645 | - | PROTEIN:hypothetical protein |
| LCRIS_01925 | LCRIS_01925 | 1937553 | 1938185 | 633 | - | PROTEIN:hypothetical protein |
| LCRIS_01926 | LCRIS_01926 | 1938297 | 1939328 | 1032 | + | PROTEIN:hypothetical protein |
| LCRIS_01927 | LCRIS_01927 | 1939508 | 1940905 | 1398 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01928 | LCRIS_01928 | 1940931 | 1941425 | 495 | - | PROTEIN:hypothetical protein |
| LCRIS_01929 | LCRIS_01929 | 1941530 | 1941889 | 360 | - | PROTEIN:hypothetical protein |
| LCRIS_01930 | LCRIS_01930 | 1942007 | 1942207 | 201 | - | PROTEIN:hypothetical protein |
| LCRIS_01931 | purB | 1942419 | 1943717 | 1299 | - | PROTEIN:adenylosuccinate lyase |
| LCRIS_01932 | purA | 1943721 | 1945010 | 1290 | - | PROTEIN:adenylosuccinate synthetase |
| LCRIS_01933 | guaC | 1945208 | 1946200 | 993 | + | PROTEIN:gmp reductase |
| LCRIS_02089 | tRNA-Arg | 1946399 | 1946472 | 74 | + | RNA:Arg tRNA |
| LCRIS_02090 | tRNA-Pro | 1946481 | 1946554 | 74 | + | RNA:Pro tRNA |
| LCRIS_01934 | LCRIS_01934 | 1946631 | 1947689 | 1059 | - | PROTEIN:surface protein |
| LCRIS_01935 | asnA | 1948123 | 1949136 | 1014 | + | PROTEIN:aspartate--ammonia ligase |
| LCRIS_01936 | LCRIS_01936 | 1949192 | 1949443 | 252 | - | PROTEIN:hypothetical protein |
| LCRIS_01937 | LCRIS_01937 | 1949843 | 1950919 | 1077 | - | PROTEIN:hypothetical protein |
| LCRIS_01938 | LCRIS_01938 | 1950924 | 1951916 | 993 | - | PROTEIN:hypothetical protein |
| LCRIS_01939 | LCRIS_01939 | 1951918 | 1952727 | 810 | - | PROTEIN:hypothetical protein |
| LCRIS_01940 | LCRIS_01940 | 1952728 | 1953096 | 369 | - | PROTEIN:hypothetical protein |
| LCRIS_01941 | LCRIS_01941 | 1953128 | 1953565 | 438 | - | PROTEIN:hypothetical protein |
| LCRIS_01942 | LCRIS_01942 | 1953558 | 1954274 | 717 | - | PROTEIN:ABC transporter, ATP-binding protein ABC2a |
| LCRIS_01943 | LCRIS_01943 | 1954271 | 1955002 | 732 | - | PROTEIN:hypothetical protein |
| LCRIS_01944 | LCRIS_01944 | 1955057 | 1955332 | 276 | - | PROTEIN:hypothetical protein |
| LCRIS_01945 | LCRIS_01945 | 1956084 | 1957481 | 1398 | - | PROTEIN:amino acid permease |
| LCRIS_01946 | LCRIS_01946 | 1957494 | 1958222 | 729 | - | PROTEIN:glutamine amidotransferase |
| LCRIS_01947 | LCRIS_01947 | 1958382 | 1958861 | 480 | - | PROTEIN:yobm related protein |
| LCRIS_01948 | LCRIS_01948 | 1958863 | 1959768 | 906 | - | PROTEIN:hypothetical protein |
| LCRIS_01949 | LCRIS_01949 | 1960000 | 1960278 | 279 | - | PROTEIN:hypothetical protein |
| LCRIS_01950 | LCRIS_01950 | 1960337 | 1961101 | 765 | - | PROTEIN:hypothetical protein |
| LCRIS_01951 | LCRIS_01951 | 1961162 | 1962535 | 1374 | - | PROTEIN:amino acid permease |
| LCRIS_01952 | cycA | 1962608 | 1963957 | 1350 | - | PROTEIN:amino acid permease |
| LCRIS_01953 | lepB2 | 1964119 | 1964751 | 633 | - | PROTEIN:signal peptidase i |
| LCRIS_01954 | clpE2 | 1964865 | 1966991 | 2127 | - | PROTEIN:ATP-dependent clp protease, ATP-binding su |
| LCRIS_01955 | LCRIS_01955 | 1967322 | 1968215 | 894 | - | PROTEIN:cell surface hydrolase |
| LCRIS_01956 | celB7 | 1968196 | 1969503 | 1308 | - | PROTEIN:cellobiose-specific pts |
| LCRIS_01957 | LCRIS_01957 | 1969597 | 1970298 | 702 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01958 | LCRIS_01958 | 1970308 | 1972848 | 2541 | + | PROTEIN:ABC transporter, permease protein |
| LCRIS_01959 | LCRIS_01959 | 1972895 | 1973131 | 237 | + | PROTEIN:hypothetical protein |
| LCRIS_01960 | LCRIS_01960 | 1973234 | 1974850 | 1617 | - | PROTEIN:polysaccharide transporter |
| LCRIS_01961 | LCRIS_01961 | 1975140 | 1976090 | 951 | + | PROTEIN:lysin |
| LCRIS_01962 | LCRIS_01962 | 1976161 | 1976877 | 717 | - | PROTEIN:hypothetical protein |
| LCRIS_01963 | LCRIS_01963 | 1977193 | 1978134 | 942 | - | PROTEIN:penicillin-binding protein |
| LCRIS_01964 | dltD | 1978134 | 1979420 | 1287 | - | PROTEIN:D-alanyl transfer protein |
| LCRIS_01965 | dltC | 1979413 | 1979652 | 240 | - | PROTEIN:D-alanine--poly(phosphoribitol) ligase sub |
| LCRIS_01966 | dltB | 1979690 | 1980928 | 1239 | - | PROTEIN:D-alanyl-lipoteichoic acid biosynthesis pr |
| LCRIS_01967 | dltA | 1980928 | 1982442 | 1515 | - | PROTEIN:D-alanine--poly(phosphoribitol) ligase sub |
| LCRIS_01968 | LCRIS_01968 | 1982458 | 1982610 | 153 | - | PROTEIN:D-ala-teichoic acid biosynthesis protein |
| LCRIS_01969 | LCRIS_01969 | 1982759 | 1983055 | 297 | - | PROTEIN:hypothetical protein |
| LCRIS_01970 | LCRIS_01970 | 1983081 | 1984217 | 1137 | - | PROTEIN:hypothetical protein |
| LCRIS_01971 | LCRIS_01971 | 1984801 | 1985271 | 471 | + | PROTEIN:cbs domain containing protein |
| LCRIS_01972 | LCRIS_01972 | 1985400 | 1987262 | 1863 | + | PROTEIN:cadmium efflux atpase |
| LCRIS_01973 | LCRIS_01973 | 1987508 | 1988677 | 1170 | + | PROTEIN:Na+/H+ antiporter |
| LCRIS_01974 | LCRIS_01974 | 1988717 | 1990276 | 1560 | - | PROTEIN:ABC transporter, atpase and permease prote |
| LCRIS_01975 | LCRIS_01975 | 1990623 | 1991297 | 675 | - | PROTEIN:hypothetical protein |
| LCRIS_01976 | LCRIS_01976 | 1991330 | 1992190 | 861 | + | PROTEIN:transcriptional regulator |
| LCRIS_01977 | LCRIS_01977 | 1992268 | 1992624 | 357 | - | PROTEIN:hypothetical protein |
| LCRIS_01978 | LCRIS_01978 | 1992860 | 1993513 | 654 | - | PROTEIN:metal-dependent membrane protease |
| LCRIS_01979 | LCRIS_01979 | 1993605 | 1994708 | 1104 | + | PROTEIN:integral hypothetical protein |
| LCRIS_01980 | LCRIS_01980 | 1994723 | 1997062 | 2340 | + | PROTEIN:ABC transporter, ATP-binding protein |
| LCRIS_01981 | ppc | 1997169 | 1999907 | 2739 | + | PROTEIN:phosphoenolpyruvate carboxylase |
| LCRIS_01982 | LCRIS_01982 | 1999999 | 2000604 | 606 | - | PROTEIN:hypothetical protein |
| LCRIS_01983 | bmpA1 | 2000910 | 2002004 | 1095 | + | PROTEIN:lipoprotein a-antigen |
| LCRIS_01984 | bmpA2 | 2002146 | 2003231 | 1086 | + | PROTEIN:lipoprotein a-antigen |
| LCRIS_01985 | LCRIS_01985 | 2003313 | 2004851 | 1539 | + | PROTEIN:sugar ABC transporter, ATP binding protein |
| LCRIS_01986 | LCRIS_01986 | 2004844 | 2005992 | 1149 | + | PROTEIN:sugar ABC transporter, permease protein |
| LCRIS_01987 | LCRIS_01987 | 2005992 | 2006948 | 957 | + | PROTEIN:sugar ABC transporter, permease protein |
| LCRIS_01988 | LCRIS_01988 | 2007052 | 2007879 | 828 | - | PROTEIN:aldo/keto reductase |
| LCRIS_01989 | nagB | 2008006 | 2008725 | 720 | + | PROTEIN:glucosamine-6-phosphate deaminase |
| LCRIS_01990 | dgk1 | 2009020 | 2009667 | 648 | + | PROTEIN:deoxyadenosine kinase |
| LCRIS_01991 | dgk2 | 2009685 | 2010371 | 687 | + | PROTEIN:deoxyguanosine kinase |
| LCRIS_01992 | LCRIS_01992 | 2010487 | 2010945 | 459 | + | PROTEIN:hypothetical protein |
| LCRIS_01993 | LCRIS_01993 | 2010996 | 2011796 | 801 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_01994 | pbuG1 | 2012025 | 2013332 | 1308 | + | PROTEIN:xanthine/uracil/vitamin c permease |
| LCRIS_01995 | LCRIS_01995 | 2013395 | 2014594 | 1200 | - | PROTEIN:atpase |
| LCRIS_01996 | LCRIS_01996 | 2014774 | 2015499 | 726 | - | PROTEIN:transcriptional regulator, merr family |
| LCRIS_01997 | pbuG2 | 2015683 | 2016993 | 1311 | + | PROTEIN:xanthine/uracil/vitamin c permease |
| LCRIS_01998 | LCRIS_01998 | 2017160 | 2017426 | 267 | + | PROTEIN:DNA-damage-inducible protein j |
| LCRIS_01999 | LCRIS_01999 | 2017416 | 2017748 | 333 | + | PROTEIN:cytosolic protein |
| LCRIS_02000 | LCRIS_02000 | 2018447 | 2018695 | 249 | + | PROTEIN:hypothetical protein |
| LCRIS_02001 | LCRIS_02001 | 2018798 | 2019343 | 546 | + | PROTEIN:hypothetical protein |
| LCRIS_02002 | LCRIS_02002 | 2019347 | 2019724 | 378 | + | PROTEIN:arac-type DNA-binding domain-containing pr |
| LCRIS_02003 | LCRIS_02003 | 2019852 | 2021066 | 1215 | + | PROTEIN:atpase |
| LCRIS_02004 | LCRIS_02004 | 2021104 | 2021877 | 774 | - | PROTEIN:had superfamily hydrolase |
| LCRIS_02005 | pbuG3 | 2022032 | 2023342 | 1311 | + | PROTEIN:xanthine/uracil/vitamin c permease |
| LCRIS_02006 | LCRIS_02006 | 2023442 | 2023684 | 243 | + | PROTEIN:hypothetical protein |
| LCRIS_02007 | LCRIS_02007 | 2023786 | 2024226 | 441 | + | PROTEIN:transcriptional regulator |
| LCRIS_02008 | LCRIS_02008 | 2024242 | 2024619 | 378 | + | PROTEIN:cupredoxin-like domain |
| LCRIS_02009 | LCRIS_02009 | 2024632 | 2024919 | 288 | + | PROTEIN:cupredoxin-like domain |
| LCRIS_02010 | copA | 2024919 | 2026859 | 1941 | + | PROTEIN:copper-translocating p-type atpase |
| LCRIS_02011 | LCRIS_02011 | 2026851 | 2027495 | 645 | - | PROTEIN:fnr/crp family transcriptional regulator |
| LCRIS_02012 | LCRIS_02012 | 2027588 | 2028142 | 555 | + | PROTEIN:hypothetical protein |
| LCRIS_02013 | LCRIS_02013 | 2028268 | 2029632 | 1365 | + | PROTEIN:major facilitator superfamily permease |
| LCRIS_02014 | LCRIS_02014 | 2029689 | 2029841 | 153 | - | PROTEIN:hypothetical protein |
| LCRIS_02015 | LCRIS_02015 | 2029902 | 2031011 | 1110 | - | PROTEIN:hypothetical protein |
| LCRIS_02016 | LCRIS_02016 | 2031507 | 2033066 | 1560 | - | PROTEIN:voltage-gated chloride protein |
| LCRIS_02017 | gnd | 2033167 | 2034570 | 1404 | - | PROTEIN:6-phosphogluconate dehydrogenase, decarbox |
| LCRIS_02018 | LCRIS_02018 | 2034743 | 2035456 | 714 | - | PROTEIN:hypothetical protein |
| LCRIS_02019 | poxL | 2035670 | 2037478 | 1809 | - | PROTEIN:pyruvate oxidase |
| LCRIS_02020 | mnmG | 2037847 | 2039751 | 1905 | - | PROTEIN:tRNA uridine 5-carboxymethylaminomethyl mo |
| LCRIS_02021 | mnmE | 2039753 | 2041138 | 1386 | - | PROTEIN:tRNA modification GTPase mnme |
| LCRIS_02022 | spoIIIJ | 2041249 | 2042124 | 876 | - | PROTEIN:stage iii sporulation protein j |
| LCRIS_02023 | rnpA | 2042126 | 2042494 | 369 | - | PROTEIN:ribonuclease p protein component |
| LCRIS_02024 | rpmH | 2042546 | 2042719 | 174 | - | PROTEIN:50S ribosomal protein l34 |