ASSP
A Program for assigning Secondary Structures in proteins
ASSP Background Information:Secondary structure Elements (SSEs) play an important role in the folding of proteins. These SSEs can be defined using repeating backbone torsion angles (φ, ψ) values or main chain-main chain (MM) N-H...O hydrogen bond (h-bond) patterns giving rise to similar values of helical parameters viz, twist, rise and vtor. Available secondary structure assignment algorithms can be broadly classified into three categories: (i) algorithms based on (φ, ψ) and MM h-bond patterns (ii) algorithms based on 3D geometry (iii) hybrid methods, which use both (i) and (ii). The correct identification of the SSEs, especially their termini is still a problem.
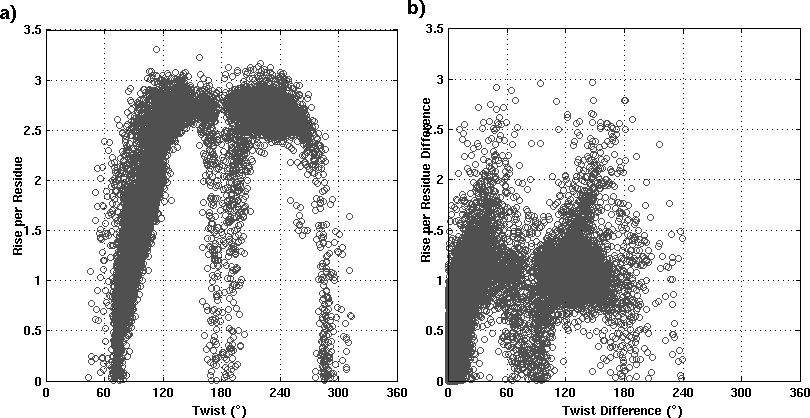
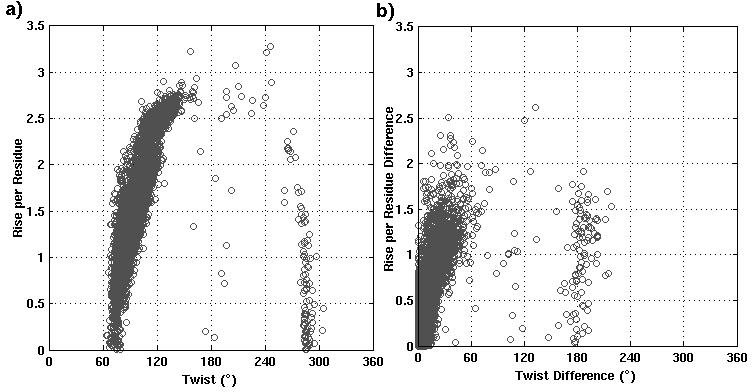
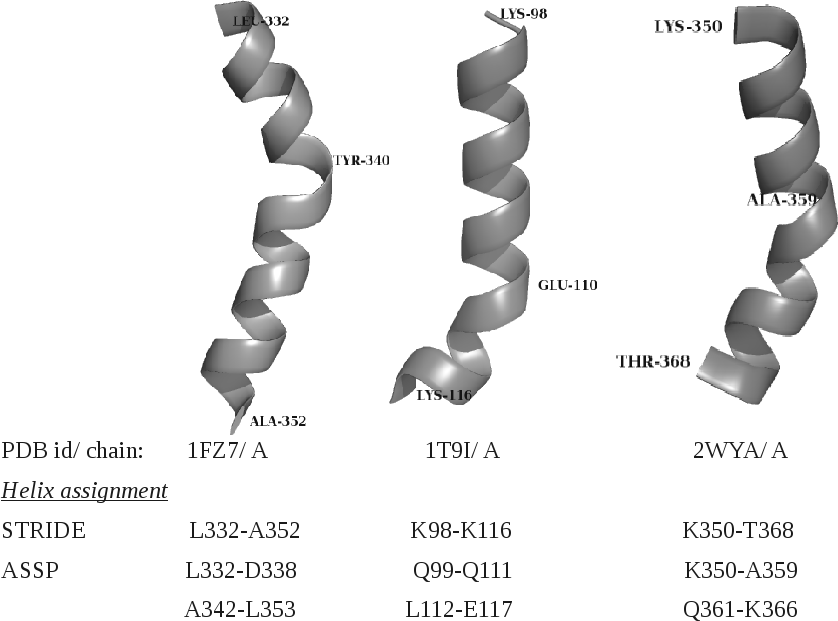
The analysis of helix records mentioned in the PDB file shows that very often the SSEs are wrongly identified (figure1). Though these discrepancies are less in DSSP/ STRIDE assignments (figure2), still the biasness of these algorithms towards the α-helices over non α-helical segments, is reported. Some of the wrong assignments is clearly illustrated in figure3. ASSP is an extension of in-house program HELANAL-Plus and uses the path traversed by the Cα atoms.
Figure 1: a) Distribution of Rise (Å) vs Twist (°) for the 71600 consecutive steps from 6052 α-helices, as mentioned in PDB file b) Distribution of absolute values of Twist difference (°) vs Rise difference (Å) between two consecutive steps in 6052 α-helices.
Figure 2: a) Distribution of Rise (Å) vs Twist (°) for the 63019 consecutive steps from 5465 commonly assigned α-helices by both DSSP and STRIDE b) Distribution of absolute values of Twist difference (°) vs Rise difference (Å) between two consecutive steps in 5465 α-helices.
Figure 3: Examples of STRIDE assigned α-helices and corresponding ASSP assignments. N & C termini are labelled along with the residue at which HELANAL-Plus shows maximum bending angle(> 30).
ASSP is written in Perl language and accepts input atomic coordinates in standard PDB format. Visual page of ASSP gives the graphical representation of SSE assignments for the input protein. Input protein will be displayed in the color format similar to the graphical representation. ASSP gives an option of comparing the result with that of two widely used SSE assignment programs,STRIDE and DSSP. It has an option of running HELANAL-Plus for the helices assigned. Helical wheel representation of the helices or any other segment of the input protein adds value to the web-server.
The web server version of ASSP is developed using PERL, a widely used scripting language & Common Gateway Interface (CGI), the standard protocol for interfacing application software with web server. Apache is used as the web server.
Copyright © 2010 [Molecular Biophysics Unit,IISC]. All rights reserved.