ASSP
A Program for assigning Secondary Structures in proteins
Description of ASSP output files:The following output files are created during each run of ASSP:
*_nh.out
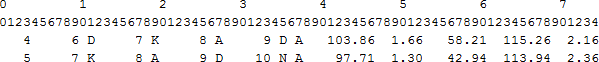
contains the unit twist, unit rise (height), virtual torsion angle, bending angle between two successive turns and radius, calculated for each set of four CA atoms in the helix.
Format of the file is as follows:
Name Start End Format Description Line Number 0 3 I4 Line number or Step number Res Number 6 9 I4 CA atom of 1st residue of the repeating unit Res Id 11 11 A1 Single letter code Res Number 14 17 I4 CA atom of 2nd residue of the repeating unit Res Id 19 19 A1 Single letter code Res Number 22 25 I4 CA atom of 3rd residue of the repeating unit Res Id 27 27 A1 Single letter code Res Number 30 33 I4 CA atom of 4th residue of the repeating unit Res Id 35 35 A1 Single letter code Chain Id 37 37 A1 Chain identifier of the repeating unit Twist 41 46 F6.2 Translation along the helical axis per repeating unit (Deg.) Height 49 52 F4.2 Rise along the helical axis per repeating unit (Ang.) Vtor 55 60 F6.2 Virtual torsional angle per repeating unit (Deg.) Bending Angle 63 68 F6.2 Bending angle b/w local helix axes of two successive turns(Deg.) Radius 71 74 F4.2 Radius of least squares circle fitted per repeating unit (Ang.) *_diff.out contains the absolute value of the differences of local parameters mentioned above.
Format of the file is as follows:
Name Start End Format Description Chain Id 0 0 A1 Chain identifier to which all five residues belong Res Num 2 5 I4 CA atom of 1st residue of the repeating unit 1 Res Id 7 7 A1 Amino acid single letter code Res Num 10 13 I4 CA atom of 4th residue of the repeating unit 2 Res Id 15 15 A1 Amino acid single letter code Twist1 18 23 F6.2 Translation along the helical axis for repeating unit 1 (Deg.) Twist2 26 31 F6.2 Translation along the helical axis for repeating unit 2 (Deg.) ΔTwist 34 39 F6.2 Absolute difference between Twist1 and Twist2 (Deg.) h1 42 45 F4.2 Rise along the helical axis for repeating unit 1 (Ang.) h2 48 51 F4.2 Rise along the helical axis for repeating unit 2 (Ang.) Δh 54 57 F4.2 Absolute difference between Height1 and Height2 (Ang.) Vtor1 60 65 F6.2 Virtual torsional angle for repeating unit 1 (Deg.) Vtor2 68 73 F6.2 Virtual torsional angle for repeating unit 2 (Deg.) ΔVtor 76 81 F6.2 Absolute difference between Vtor1 and Vtor2 (Deg.) Res Num 84 87 I4 CA atom of 2nd residue of the repeating unit 1 Res Id 89 89 A1 Amino acid single letter code Res Num 92 95 I4 CA atom of 3rd residue of the repeating unit 2 Res Id 97 97 A1 Amino acid single letter code
*_cont.out Two successive repeating units are part of continuous stretch, if and only if absolute value of (ΔTwist, Δh, ΔVtor) < (35˚, 1.1Å, 50˚). Contains the steps of a continuous stretch with step wise helix type (explained below) assignment. Assignments are added in the begining of the corresponding step in *_nh.out. Added assignments can be A, G, I, P, S, a, g, i or U (explained below). This file also contains the average value of twist and rise of each continuous stretch (Format is described bellow).
Format of the file is as follows:
Name Start End Format Description Assignment1 0 0 A1 Twist, Rise and Radius satisfying the criteria of either α helix or PPII (can have A, a, S, P, U) Assignment2 2 2 A1 Twist, Rise and Radius satisfying the criteria of 310 helix (can have G, g, U) Assignment3 4 4 A1 Twist, Rise and Radius satisfying the criteria of π helix (can have I, i, U) Description of assignments explained above:
Helix type class number Description Helix type class number Description A 1 Right-handed α helix a 6 Left-handed α helix G 5 Right-handed 310 helix g 12 Left-handed 310 helix I 3 Right-handed π helix i 11 Left-handed π helix P 10 PPII helix U - Undefined S 0 Strands (Extended Configuration) - - - Description of the record consisting of avg. Twist and avg. Rise:
Name Start End Format Description File Name 0 6 A6 Name of the input file (without the extension) Cont.Num. 10 13 I4 Continuous stretch number Res ID 15 17 A3 Starting residue (3 Letter code) Chain Id 19 19 A1 Chain Identifier Res Num 21 24 I4 Residue Number (same as in PDB) Res ID 27 29 A3 End Residue (3 Letter code) Chain Id 31 31 A1 Chain Identifier Res Num 33 36 I4 Residue Number (same as in PDB) Avg. Twist 47 52 F6.2 Average unit twist of the helix (Deg.) Avg. Height 55 58 F4.2 Average unit height of the helix (Ang.) *_assp.out contains the secondary structure assignment by ASSP in PDB "HELIX record" format.
Format of the file is as follows:
Name Start End Format Description SS Type 0 9 A10 Secondary Structure Type SS Num 11 13 I3 Serial Number of Secondary Structure Res ID 15 17 A3 Starting residue (3 Letter code) Chain Id 19 19 A1 Chain Identifier Res Num 21 24 I4 Residue Number (same as in PDB) Res ID 27 29 A3 End Residue (3 Letter code) Chain Id 31 31 A1 Chain Identifier Res Num 33 36 I4 Residue Number (same as in PDB) SS Identifier 38 39 I2 Secondary Structure Type Length 73 75 I3 Length of the Secondary Structure *_tor.out file contains the phi-psi torsional angle record for each residue.

Name Start End Format Description Res Id 1 3 A3 3 Letter code of the residue Chain Id 7 7 A1 Chain Identifier of the residue Res Num 10 13 I4 Residue Number (same as in PDB) Phi Angle 17 23 F7.2 Phi Torsional Angle Psi Angle 27 33 F7.2 Psi Torsional Angle
Copyright © 2010 [Molecular Biophysics Unit,IISC]. All rights reserved.